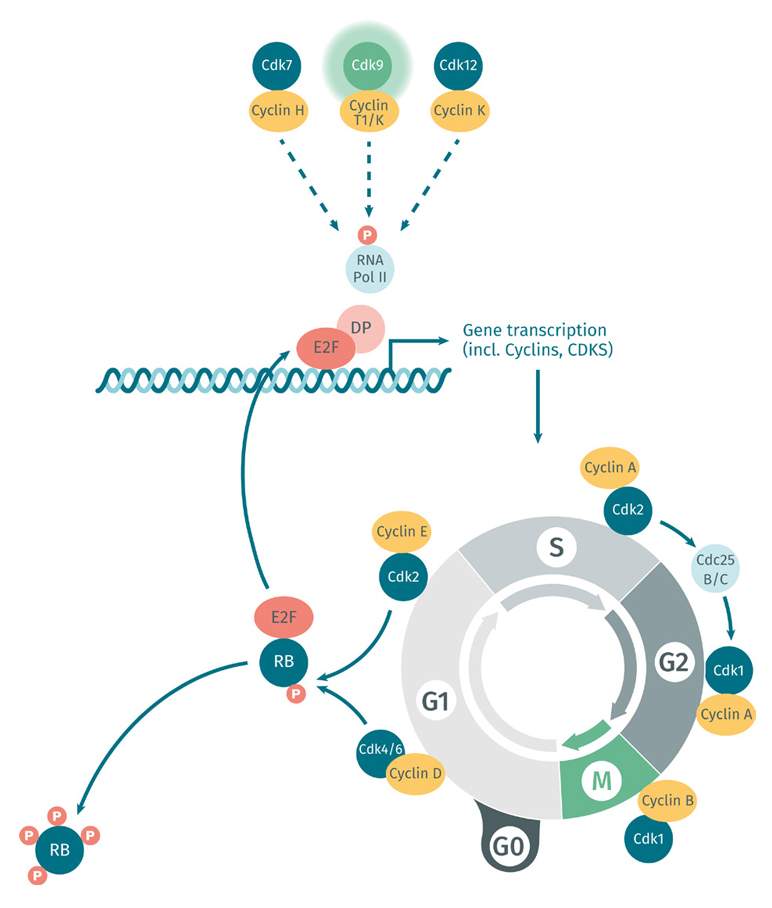
Cancer cells achieving immortality
One of the reasons cancer cells are so difficult to treat is their remarkable ability to achieve immortality. Unlike normal cells, which have a limited lifespan, cancer cells can divide indefinitely, enabling them to grow and spread throughout the body. This is due to various genetic mutations that allow cancer cells to circumvent the normal regulatory mechanisms that keep cell growth and proliferation in check. Scientists are continuing to study these mutations in the hope of finding new ways to target and eliminate resilient cancer cells for oncology drug development.
One of the reasons cancer cells are so difficult to treat is their remarkable ability to achieve immortality. Unlike normal cells, which have a limited lifespan, cancer cells can divide indefinitely, enabling them to grow and spread throughout the body. This is due to various genetic mutations that allow cancer cells to circumvent the normal regulatory mechanisms that keep cell growth and proliferation in check. Scientists are continuing to study these mutations in the hope of finding new ways to target and eliminate resilient cancer cells for oncology drug development.
Resisting cell death/cell survival promotion
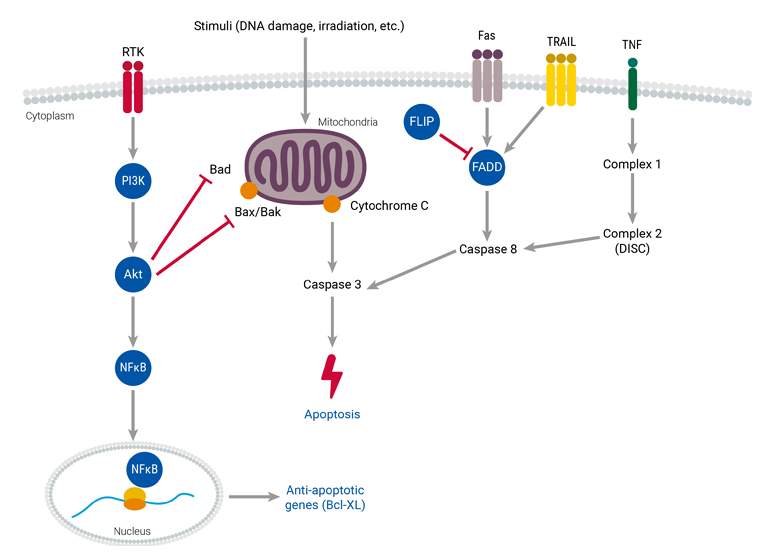
Understanding the mechanisms underlying cancer cells’ resistance to cell death is important for developing effective treatments. Cancer cells have the ability to evade apoptosis, the natural process of programmed cell death that helps maintain the health of an organism. This evasion can occur through alterations in apoptosis detection and signaling mechanisms, as well as defects in the downstream signaling pathways and related proteins. To better understand how cancer cells evade cell death, it is important to investigate various apoptosis pathways, such as the activation of death receptors or involvement of proteins like caspase-8, RIP kinases, Bcl-2, and p53.
Genome instability and mutation
A variety of factors influence the quality of cancer cells, including competition for resources with neighboring cells, the evasion of immune responses, and self-suppression. These factors can lead to genomic instability, which can increase the likelihood of mutations occurring in future cells. Environmental factors and DNA maintenance defects can further increase this risk, while epigenetic modifications can also contribute to cancer cell growth. It is important to note that not all cancer cells are equal; they evolve in response to selective pressures driven by the accumulation of genetic mutations over time. There are several key proteins involved in maintaining genomic stability and preventing DNA damage. These include DNA-dependent protein kinase, BRCA1 and BRCA2, Chk1 and Chk2, and p53. These proteins play critical roles in mitigating factors that could trigger DNA damage and increase the risk of malignancy.
Tumor Invasion and Metastasis
The ability of cancer cells to invade nearby tissues and spread to other parts of the body is known as tumor invasion and metastasis. This dangerous aspect of cancer can lead to the formation of new tumors in distant organs and tissues. A crucial factor in this process is the cancer cells’ ability to achieve immortality, enabling them to continuously divide and spread, even after leaving the original tumor site. Researchers are working to better understand the mechanisms behind tumor invasion and metastasis in order to develop more effective treatments for advanced stages of cancer.
The ability of cancer cells to invade nearby tissues and spread to other parts of the body is known as tumor invasion and metastasis. This dangerous aspect of cancer can lead to the formation of new tumors in distant organs and tissues. A crucial factor in this process is the cancer cells’ ability to achieve immortality, enabling them to continuously divide and spread, even after leaving the original tumor site. Researchers are working to better understand the mechanisms behind tumor invasion and metastasis in order to develop more effective treatments for advanced stages of cancer.
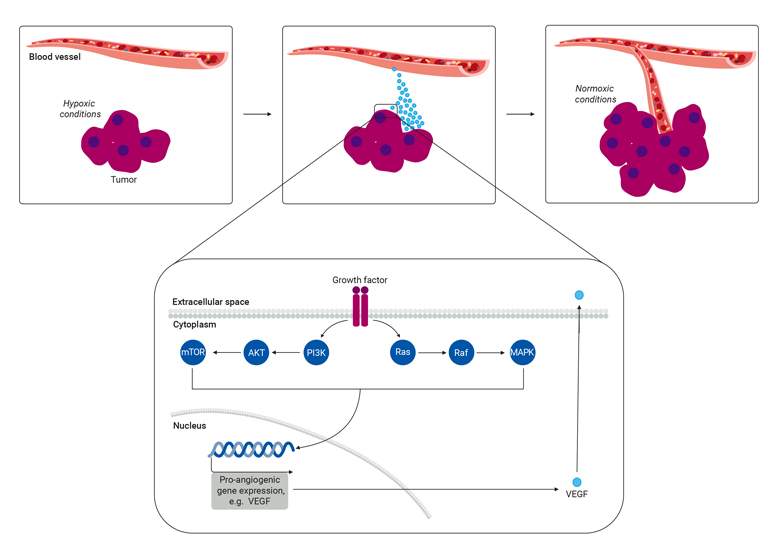
Angiogenesis
Tumor angiogenesis is a continuous process triggered by extracellular signals such as hypoxia or growth factors. The primary driver of this activity is hypoxia-inducible factor (HIF), which upregulates angiogenic growth factors such as vascular endothelial growth factors (VEGF). VEGF and HIF are important signaling proteins that attract endothelial cells to the tumor mass and stimulate new blood vessels or induce the growth of pre-existing ones. The regulation of angiogenesis relies on the PI3K/AKT and MAPK signaling pathways, which respond to growth factors by increasing HIF and VEGF expression. It is important to understand these pathways as researchers explore innovative strategies to target immune checkpoints as a potential avenue for treating cancer.
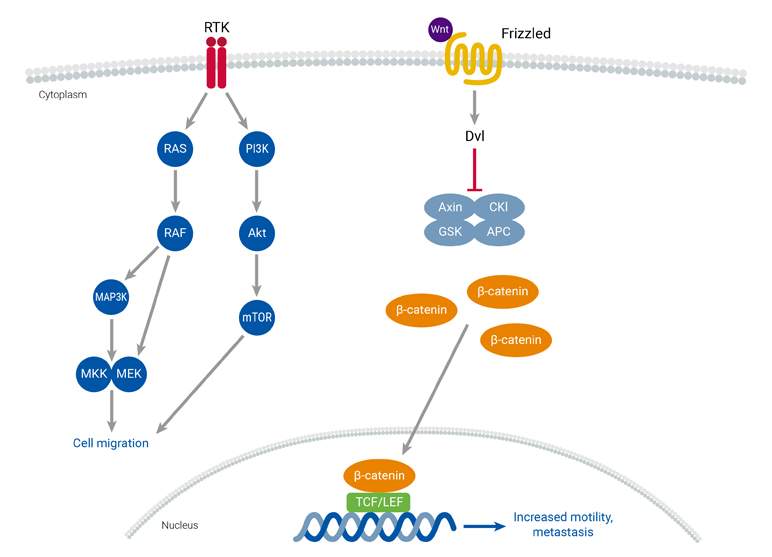
Cell Migration
Cancer development and metastasis are influenced by increased cell migration, a process that can be triggered by genetic and epigenetic changes, along with the dysregulation of cell migration signaling pathways. Metastasis involves several key events, including epithelial-mesenchymal transition (EMT), the formation of new blood vessels (tumor neoangiogenesis), and the spread of malignancy through blood vessels to distant tissues and organs. Two key pathways involved in controlling cell migration are the Wnt and receptor tyrosine kinase (RTK) pathways. Both are frequently overactivated in solid tumors and metastasis. RTKs are cell surface receptors responsible for mediating signaling pathways involved in cell migration. Mutations that affect RTKs can result in increased cell migration. Additionally, pharmacological inhibition of the PI3K-Akt signaling pathway in cells with hyperactivated Wnt signaling has been shown to increase metastasis.
Epithelial-mesenchymal transition
During biological processes and cancer progression, epithelial cells undergo a transformative change known as epithelial-mesenchymal transition (EMT). This process entails changes in epithelial cell polarity and transitions from an epithelial to a mesenchymal phenotype. These alterations, which are regulated by highly conserved pathways, enable cancer cells to be more mobile and invasive. Cancer cells that undergo EMT secrete cytokines such as TGF-β, which is the primary inducer of EMT. Additionally, the activation of EMT transcription factors leads to the downregulation of specific genes that encode proteins involved in the formation of adherens, tight junctions, and desmosomes, and the maintenance of apical-basal cell polarity. The mesenchymal phenotype that arises from EMT also promotes cell migration, which increases cancer motility and facilitates their invasion into neighboring tissues. This transition is particularly relevant in the context of metastasis, as one of the first steps involves the invasion of cancer cells into the extracellular matrix. Thus, EMT serves as a hallmark process that allows cells to migrate and invade, a pivotal step in metastatic progression.
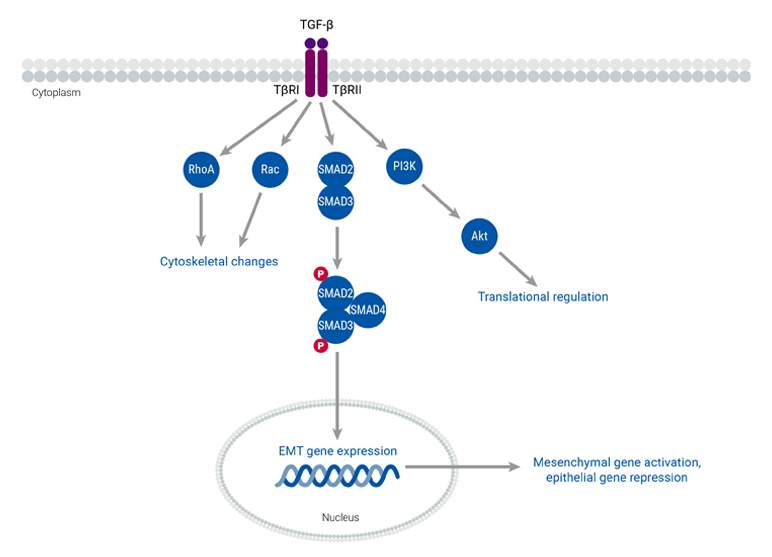
Composition of the tumor stromal microenvironment
The tumor stromal microenvironment is made up of various cell types that surround and support cancer cells, including fibroblasts, immune cells, and blood vessels. These cells communicate with each other and the cancer cells, influencing tumor growth and response to therapy. Understanding the composition of the tumor stromal microenvironment is important for developing new cancer treatments that target these interactions.
The tumor stromal microenvironment is made up of various cell types that surround and support cancer cells, including fibroblasts, immune cells, and blood vessels. These cells communicate with each other and the cancer cells, influencing tumor growth and response to therapy. Understanding the composition of the tumor stromal microenvironment is important for developing new cancer treatments that target these interactions.
ECM
The extracellular matrix (ECM) plays a crucial role in the tumor microenvironment by providing structural support to cells and regulating their behavior. Changes in the ECM, such as dysregulation of cell adhesion, migration, and differentiation, can lead to the development and metastasis of cancer. The major components of the ECM include collagen, proteoglycans, laminin, and fibronectin, which interact with cell surface receptors and signaling pathways to mediate cellular processes. Abnormal ECM dynamics compromise the integrity of the basement membrane and promote epithelial-mesenchymal transition, enabling cancer cells to infiltrate surrounding tissues. The dysregulated ECM also promotes tumor-associated angiogenesis and inflammation, creating a tumorigenic microenvironment that facilitates cancer cell invasion and metastasis. Numerous molecular networks and extracellular molecules regulate these processes, including ECM molecules, ECM receptors, and growth factors.
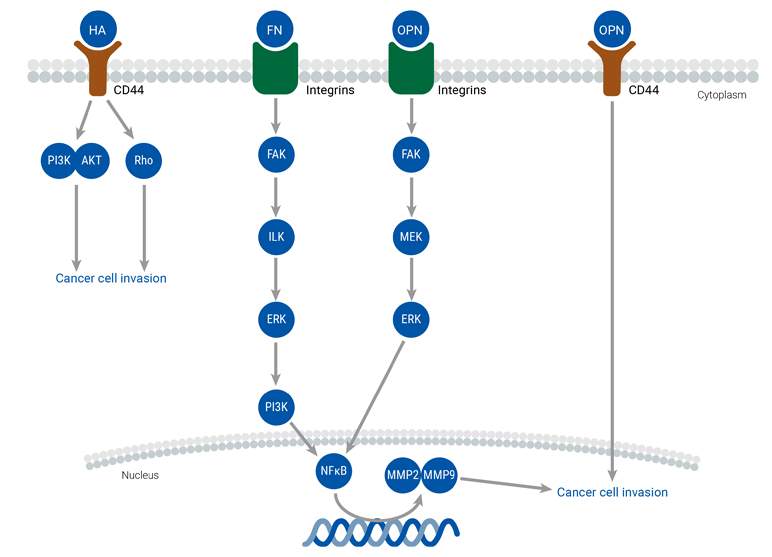
TME
The relationship between cancer cells and the tumor microenvironment (TME) is complex and constantly evolving. The TME is made up of various components, such as the extracellular matrix (ECM), endothelial cells, adipose cells, tumor-infiltrating immune cells, and cancer-associated fibroblasts (CAFs), among others. Notably, CAFs play a significant role in facilitating communication between cancer cells and the TME, ultimately promoting tumor development and migration. Stromal cells present in various adult tissues also contribute to the function and development of the TME, while the presence of immune cells can either suppress or promote tumor growth. The TME regulates cancer metastasis by releasing factors that trigger signaling pathways, leading to EMT, migration, and invasion.


Role of Inflammation and Immunity
One of the immune system’s essential functions is to help defend the body against abnormal malignancies by recognizing and destroying cells that have achieved immortality. However, tumor cells employ various mechanisms to elude the immune system, including evolving to evade immune cell destruction and secreting substances such as cytokines to promote inflammation.
One of the immune system’s essential functions is to help defend the body against abnormal malignancies by recognizing and destroying cells that have achieved immortality. However, tumor cells employ various mechanisms to elude the immune system, including evolving to evade immune cell destruction and secreting substances such as cytokines to promote inflammation.
Within immune cells, regulatory mechanisms, such as immune checkpoints which help the immune system distinguish between self and non-self and to avoid harming healthy tissues, become dysregulated. Instead of targeting cancer cells, these immune cells start secreting substances that promote the growth and spread of tumors. This process involves several molecules and signaling pathways, such as NF-κB, inflammasome signaling, immune checkpoint signaling, and markers for tumor-infiltrating immune cells. While this complexity poses a challenge, researchers are tirelessly working to discover ways to disrupt this process and enable the immune system to effectively combat cancer.
Learn more on our dedicated immuno-oncology page.

Epigenetic modifications in cancer
Epigenetic changes are critical in shaping cancer progression by influencing key cellular processes such as cell proliferation, apoptosis, invasion, and senescence. These modifications, which include DNA methylation, histone modification, and regulation through non-coding RNA, collectively play a pivotal role in tumor development and progression by altering gene expression without changing the underlying DNA sequence.
Epigenetic changes are critical in shaping cancer progression by influencing key cellular processes such as cell proliferation, apoptosis, invasion, and senescence. These modifications, which include DNA methylation, histone modification, and regulation through non-coding RNA, collectively play a pivotal role in tumor development and progression by altering gene expression without changing the underlying DNA sequence.
This is especially important to the development of tumors as our genome is equipped with a set of pro- and anti-tumorigenic genes that usually balance each other and ensure tissue homeostasis. In tumors however, the expression level of these genes is very often unbalanced, with anti-tumorigenic ones becoming repressed while the pro-tumorigenic drivers are overexpressed.
Types of Epigenetic Modifications:
- DNA Methylation: Involves the addition of methyl groups to DNA, typically suppressing gene expression and contributing to the silencing of tumor suppressor genes in cancer.
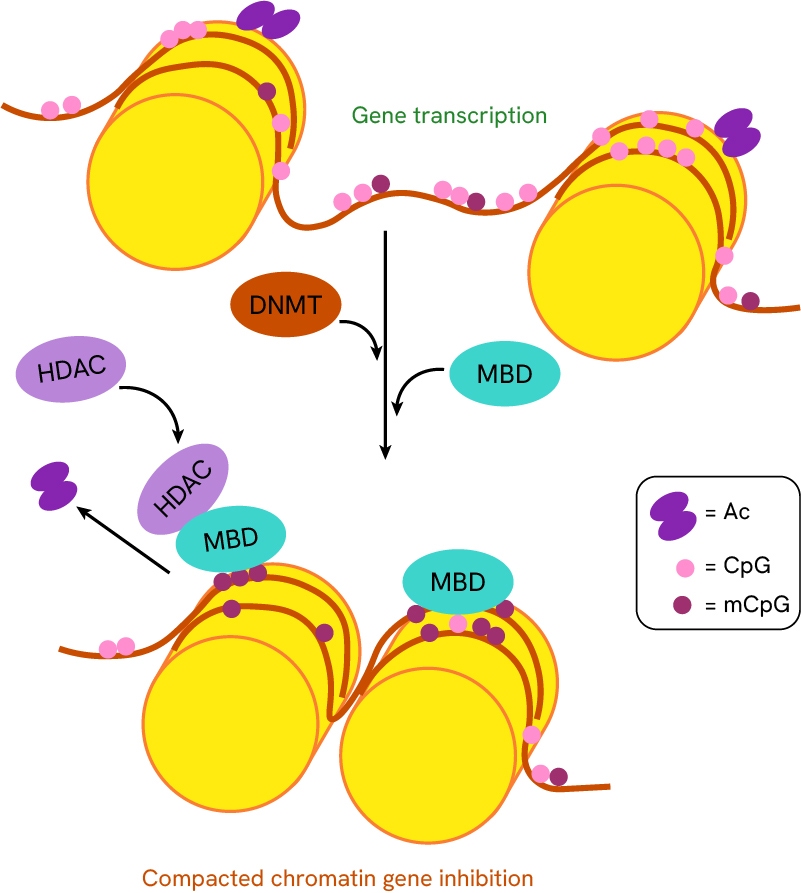
The addition of methyl groups to DNA by DNA methyltransferases promotes methyl-CpG-binding domains recruitment, which in turn recruits histone deacetylases (HDACs). The combined DNA-methylation and histone deacetylation results in an increased compact chromatin state of DNA.
- Histone Modification: Alters the structure of chromatin through the addition or removal of chemical groups, impacting how tightly DNA is packaged and influencing gene accessibility.
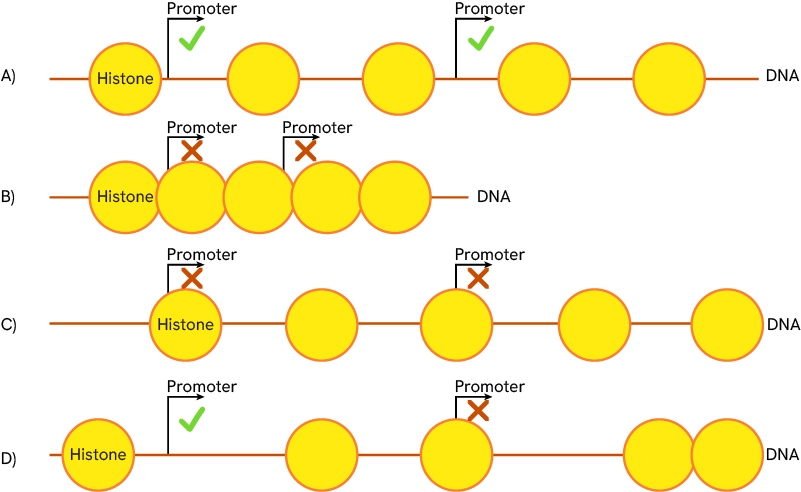
The acetylation/methylation state of histone nucleosomes is key to regulating DNA accessibility as it determines the position of histones and DNA compaction. A) Regular state of compaction, histones are spaced, and gene promoters are accessible. B) Compacted chromatin state, histones are tightly grouped, and promoters are inaccessible. C) and D) different histone sliding scenarios are possible. All histones of a region can slide together (C) or only some of them may do so (D). Promoter accessibility varies according to the histone positions.
- Non-coding RNA: Non-coding RNAs, such as microRNAs, regulate gene expression post-transcriptionally, playing a key role in modulating oncogenes and tumor suppressor genes.
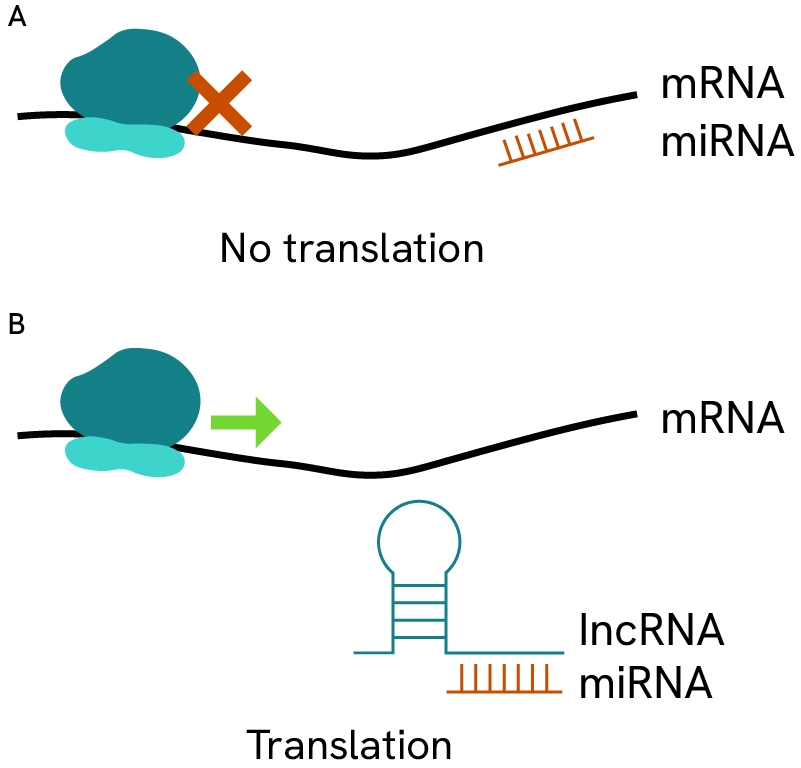
Small non-coding miRNAs regulate gene expression via inhibition/sequestration/degradation of coding mRNA transcripts. By binding to specific sequence on mRNA, these small strands can cover or uncover promoter regions, block ribosomes, or promote secondary folded structures in mRNAs that render them inaccessible for translation. Depending on the affected genes, these regulations can play protective or deleterious roles in many types of cancer.
To learn more about epigenetic modifications and their role in cancer therapy, explore our white paper, and discover our range of reagents designed to support your research in this critical area.
Featured resources


Filters
1 - 25 of 486 Products and Services
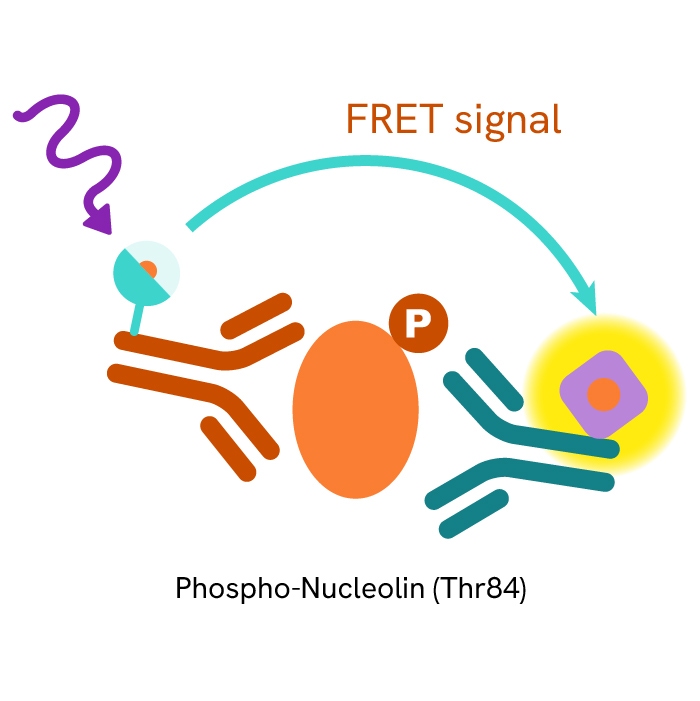
This HTRF kit allows for the cell-based quantitative detection of Nucleolin when phosphorylated at Thr84.
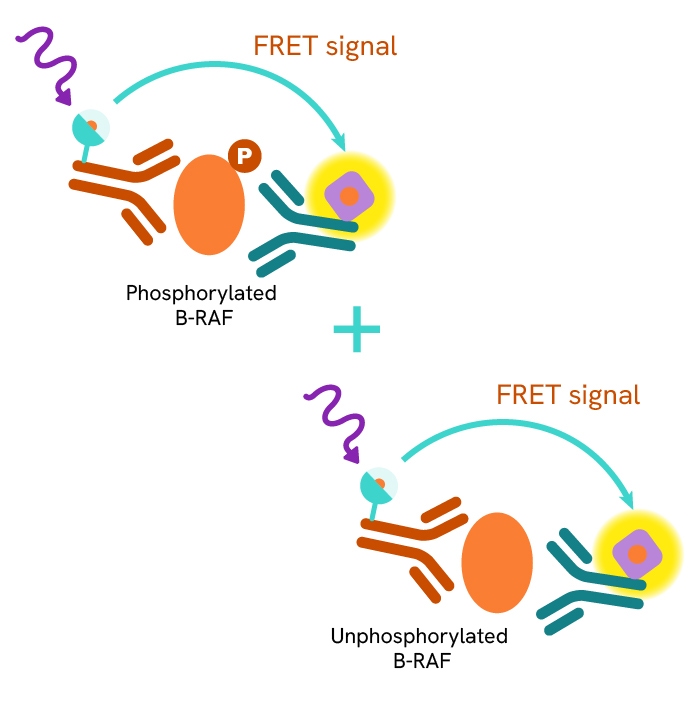
This HTRF kit allows for the cell-based quantitative detection of total B-RAF.
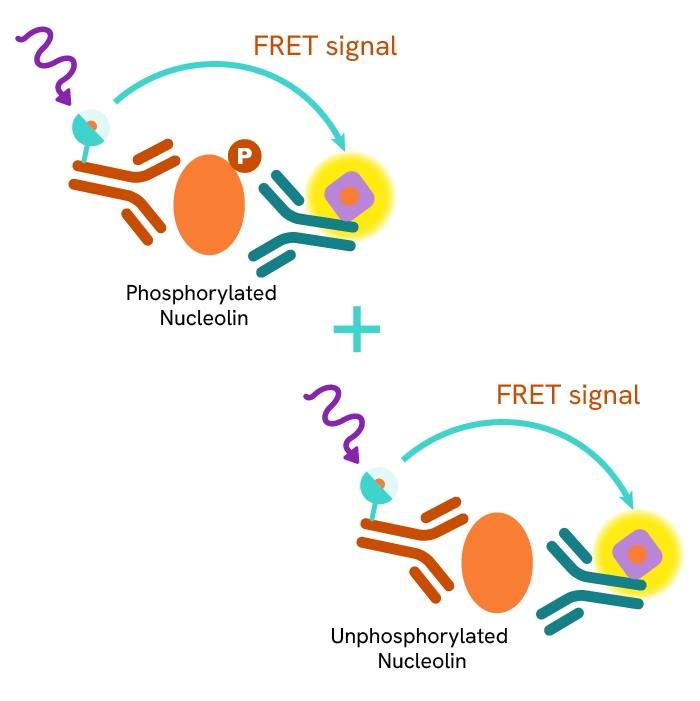
This HTRF kit allows for the cell-based quantitative detection of total Nucleolin.
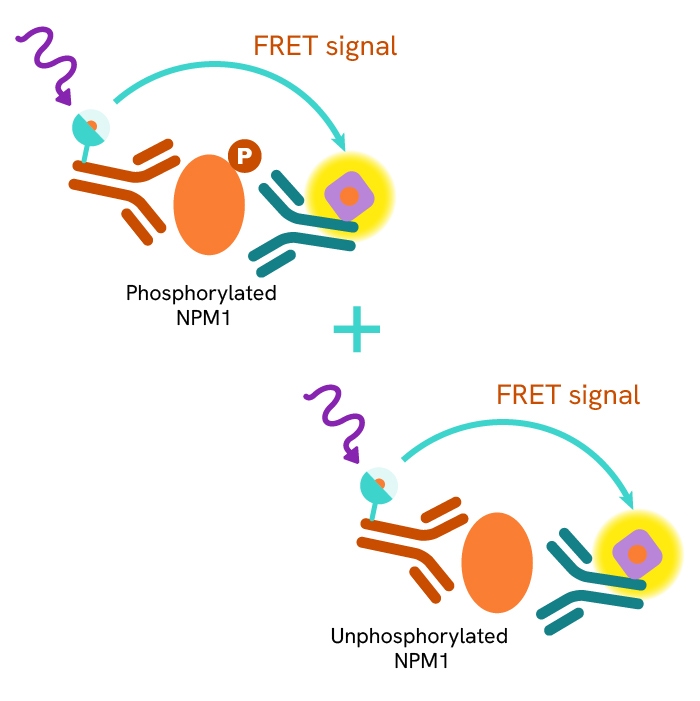
This HTRF kit allows for the cell-based quantitative detection of Total NPM1.
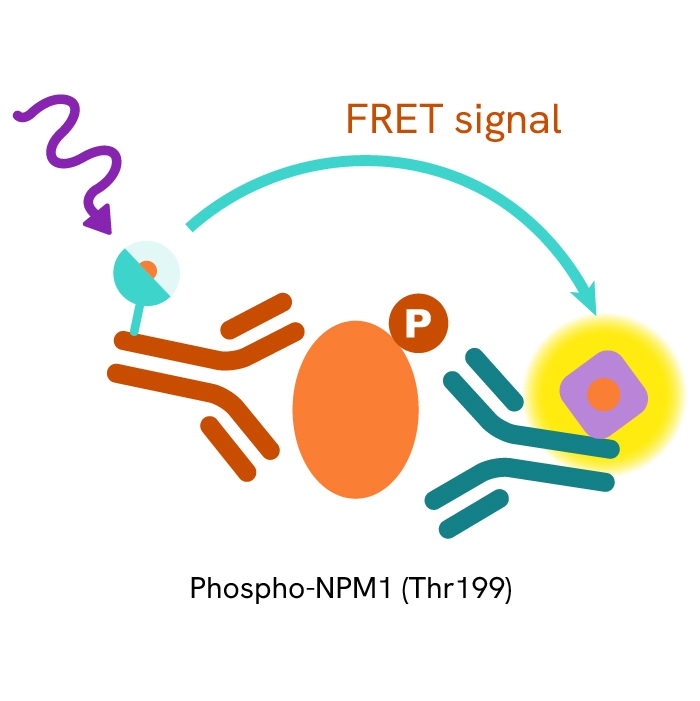
This HTRF kit allows for the cell-based quantitative detection of NPM1 when phosphorylated at Thr199.
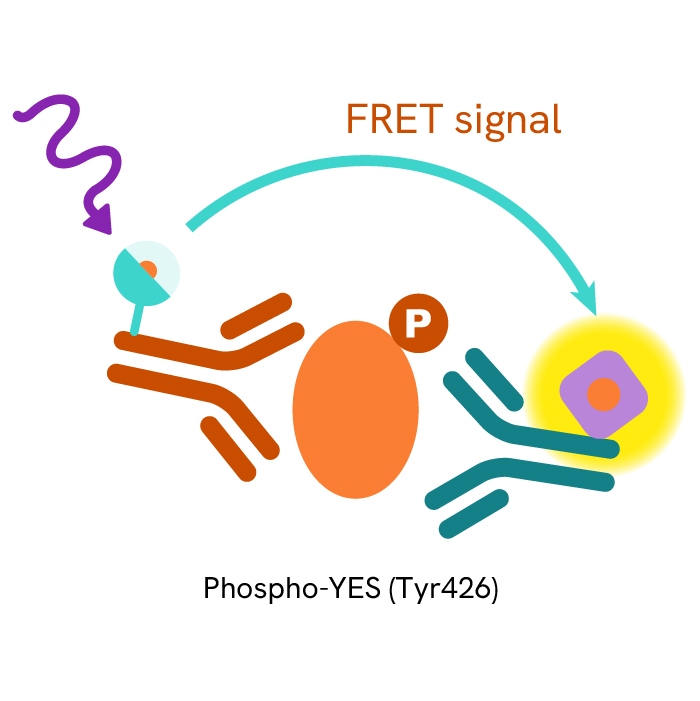
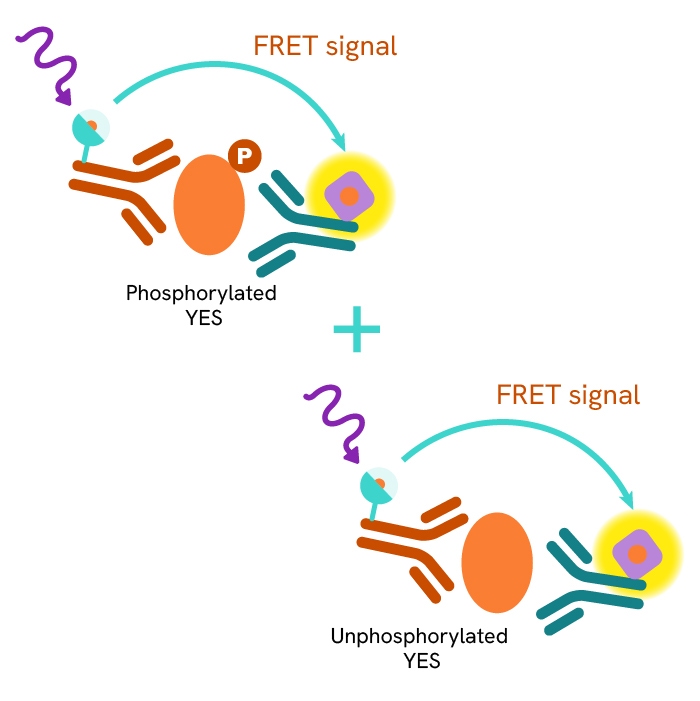
This HTRF kit allows for the cell-based quantitative detection of YES when phosphorylated at Tyr426.
This HTRF kit allows for the cell-based quantitative detection of Total YES.
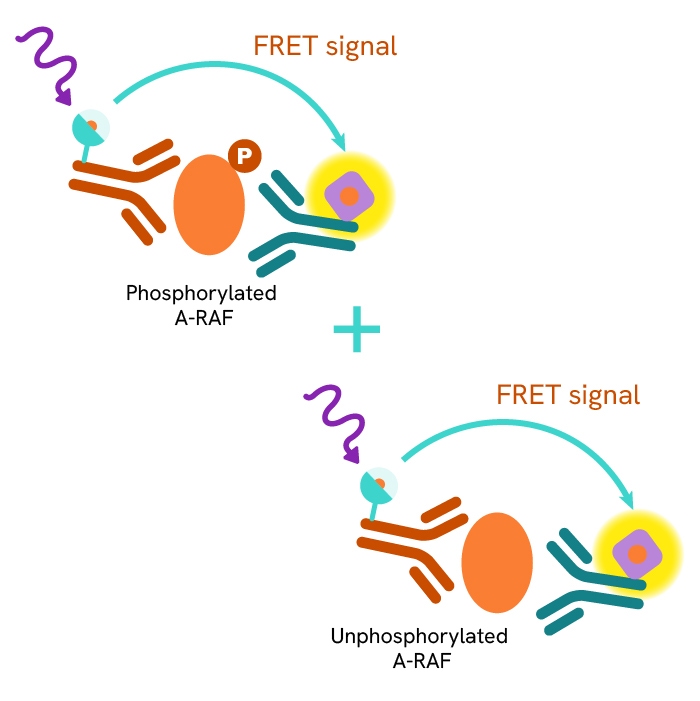
This HTRF kit allows for the cell-based quantitative detection of total A-RAF.
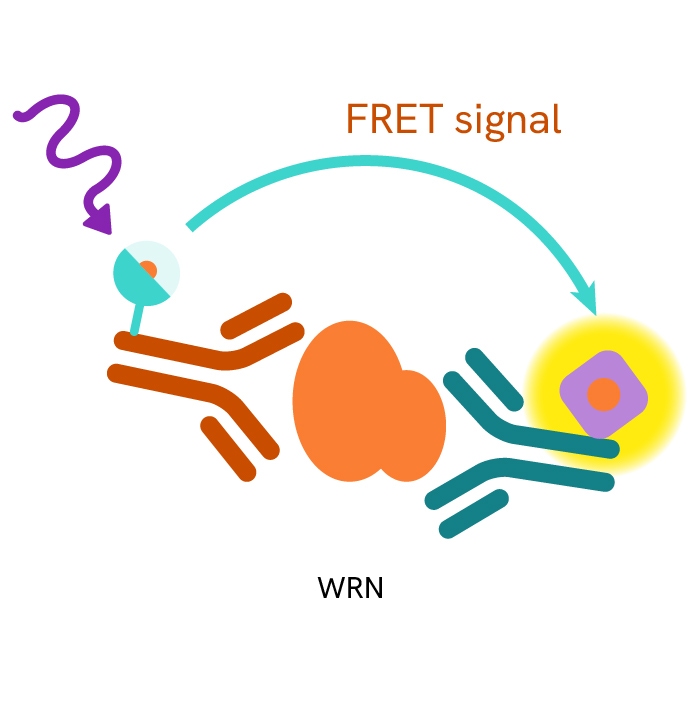
This HTRF kit allows for the cell-based quantitative detection of WRN.
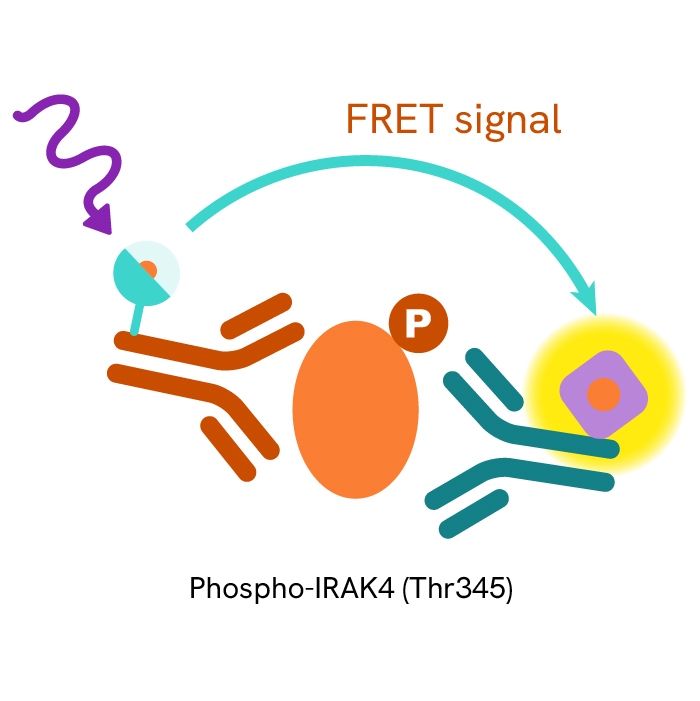
This HTRF kit allows for the cell-based quantitative detection of IRAK4 when phosphorylated at Thr345.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Phospho-Rb (Thr821) assay is a sandwich immunoassay for quantitative detection of phospho-Rb in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human Total Progesterone Receptor assay is a sandwich immunoassay for quantitative detection of total Progesterone Receptor in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Phospho-PRAS40 (Thr246) assay is a sandwich immunoassay for quantitative detection of phospho-PRAS40 in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human Total p16 INK4A assay is a sandwich immunoassay for quantitative detection of total p16 INK4A in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Total MEK2 assay is a sandwich immunoassay for quantitative detection of total MEK2 in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Phospho-MEK2 (Ser217/221) assay is a sandwich immunoassay for quantitative detection of phospho-MEK2 in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human Total p21 CiP1 assay is a sandwich immunoassay for quantitative detection of total p21 CiP1 in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Total PRAS40 assay is a sandwich immunoassay for quantitative detection of total PRAS40 in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Phospho-Rb (Thr826) assay is a sandwich immunoassay for quantitative detection of phospho-Rb in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Total BAD assay is a sandwich immunoassay for quantitative detection of total BAD in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Phospho-BAD (Ser136) assay is a sandwich immunoassay for quantitative detection of phospho-BAD in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Cleaved PARP1 D214 assay is a sandwich immunoassay for quantitative detection of cleaved PARP1 D214 in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human Phospho-Progesterone Receptor (Ser190) assay is a sandwich immunoassay for quantitative detection of phospho-Progesterone Receptor in cellular lysates using Alpha Technology.
The AlphaLISA™ SureFire® Ultra™ Human and Mouse Phospho-BAD (Ser112) assay is a sandwich immunoassay for quantitative detection of phospho-BAD in cellular lysates using Alpha Technology.
This HTRF kit allows the cell-based quantitative detection of Total CK1a.