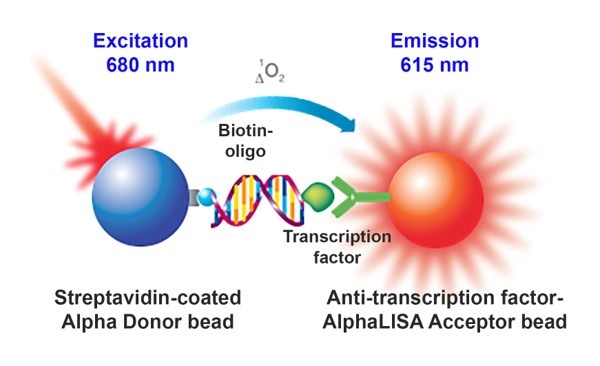
Overview
Alpha technology can be used to replace electrophoretic mobility shift assays (EMSA) to monitor DNA-protein interactions. Herein, we describe a homogeneous Alpha assay detecting a transcription factor (TF) binding specifically to its cognate DNA element. This novel AlphaLISA™ application represents a major improvement over standard EMSA assays because it eliminates the need of running gels and using radioactive materials. Moreover, in addition to its increased assay speed of execution, Alpha allows for the reduction of reagent consumption owing to its high sensitivity.
In this assay, a DNA sequence is biotinylated and captured by streptavidin-coated Donor beads while a specific antibody to the transcription factor of interest is captured by an anti-transcription factor coated AlphaLISA Acceptor bead. When the beads are brought in proximity, by the presence of the nuclear extracted transcription factors interacting with the biotinylated DNA, laser excited Donor beads will transfer energy to the Acceptor bead resulting in the emission of light.
This set-up, as depicted in Figure 1, generates a signal increase.

What do I need to run this assay?
Required reagents available from Revvity:
- A streptavidin Alpha Donor bead (see the list of available beads in Alpha reagents and catalog numbers below)
- An appropriate AlphaScreen™ or AlphaLISA Acceptor bead (see the list of available beads in Alpha reagents and catalog numbers below)
- Microplates - We recommend our 96-well 1/2 AreaPlates or our 384-well white OptiPlates™. Also see Microplate selection.
- TopSeal™-A adhesive plate seal, catalog number 6050185 for incubations
Required reagents available from various suppliers:
- Biotinylated and non-biotinylated DNA sequences (consensus and mutated) specific to the transcription factor of interest
- Nuclear extract preparation
- Specific antibody against the transcription factor Instrumentation/equipment:
- A plate reader capable of reading Alpha assays (see Instrument options on the Alpha main page)
Alpha products and catalog numbers
View our listing of relevant Alpha products with catalog numbers.
Assay protocol
In a 384-well Optiplate:
- Add 5 μL nuclear extract
- Add 5 μL assay buffer or non-biotinylated DNA sequence (for a competitive assay)
- Add 5 µL biotinylated oligonucleotides.
- Incubate the reaction at 23 °C
- Add 5 µL Ab pre-mixed with AlphaLISA beads*
- Incubate the reaction at 23 °C
- Add 5 µL Streptavidin-coated Donor beads
- Incubate the reaction in the dark at 23 °C.
-
Read using an EnVision™ Multilabel Plate Reader (Revvity)
(step 5) The antibody could be directly conjugated to the AlphaLISA acceptor beads or could be pre-incubated with AlphaLISA beads available in the ToolBox portfolio (such as Protein A, Protein G or other) as is the case for results presented below.
Assay buffer: 25 mM Hepes, 200 mM NaCl, and 0.1% Tween (pH7.4)
Assay optimizations
The data below are shown as proof-of-concept and are intended to provide guidance on how you can optimize any Alpha EMSA. Both Sp1 and HNF1 are transcription factors.
Optimal concentration of biotinylated oligonucleotide was first determined using fixed Ab and lysate concentration. Both beads were used at 20 µg/mL. Maximum response was observed using (A) 10 nM and (B) 30 nM of oligonucleotides, with signal to background ratio (S/B) values of 45.4 and 5.9, respectively.
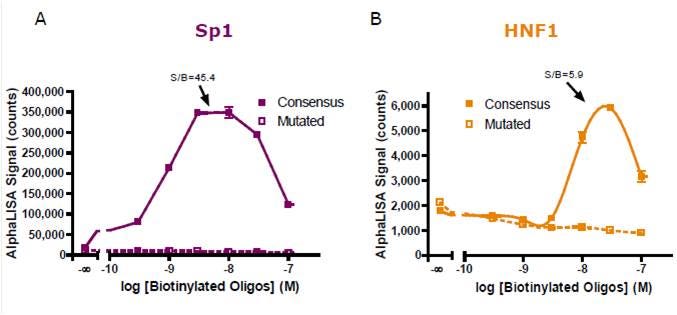
Figure 2. Biotinylated oligonucleotide titration for (A) Sp1 and (B) HNF1 detection.
An antibody titration experiment was next performed using optimal oligonucleotide concentration. The greatest assay windows were obtained with 10 and 1 nM of antibody for (A) Sp1 and (B) HNF1 detections, respectively.
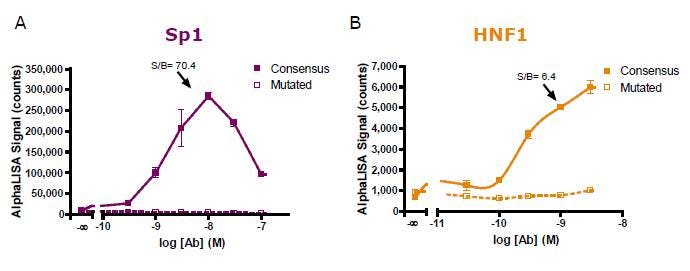
Figure 3. Antibody titration for (A) Sp1 and (B) HNF1 detection.
Using optimized reagent concentrations, time course experiments for each incubation period were then carried out and the results produced for the detection step are presented in Figure 4. A final incubation of 60 minutes was chosen for further experiment assessments even though the S/B value could be improved for Sp1 detection by incubating the reaction for a longer period of time. Note that plates could also be read after overnight incubation.
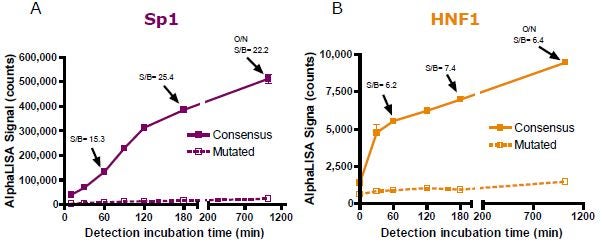
Figure 4. Detection time course experiment for (A) Sp1 and (B) HNF1 detection.
A lysate titration experiment was then conducted for both (A) Sp1 and (B) HNF1 detection using optimal assay conditions. As low as 1 µg/well of non-stimulated Hep G2 nuclear extracts permit the generation of S/B values of (A) 50.4 and (B) 2.6. However, a wider assay window (6.4) for the HNF1 assay could be generated by increasing the amount of lysate per well to 10 µg.
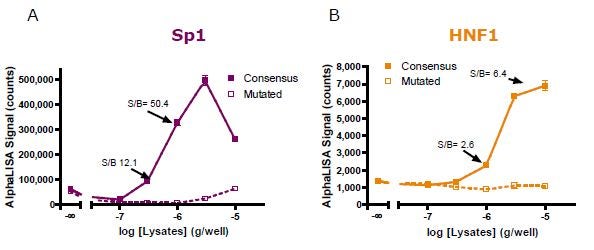
Figure 5. Nuclear extract titration for (A) Sp1 and (B) HNF1 detection.
Assay robustness was then demonstrated by generating Z’ values of (A) 0.76 and (B) 0.54 for Sp1 and HNF1 detection assays, respectively. The assay was performed using optimal assay conditions in the presence of (A) 1 µg/well and (B) 5 µg/well of Hep G2 nuclear extract.
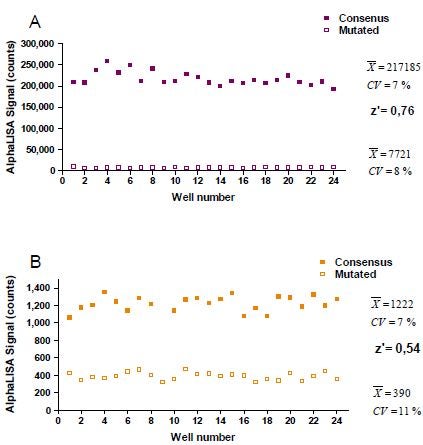
Figure 6. Assay variability experiment for Sp1 and HNF1 with Z' values.
The assay specificity was then demonstrated for Sp1 using untagged wild-type and non-specific DNA sequences as competitor for the Sp1 detection assay. A 2-log difference in potency was observed between the consensus and non-specific sequences with IC50 values of 2.3 and 241 nM, respectively.
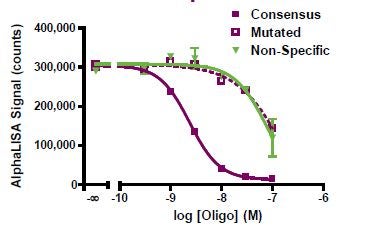
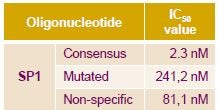
Displacement assay
For detailed information on assay design and development, view our Create your own Alpha assay page.
Conclusion
We have successfully demonstrated the application of a homogenous AlphaLISA assay to replace EMSA in the monitoring of DNA-protein interaction from nuclear extract samples. This novel AlphaLISA detection assay represents a major improvement over EMSA
- Non-radiometric assay
- Uses less DNA binding element
- Easier and faster to execute
- Greater throughput
- No need for running a gel
- Higher sensitivity
- Assay robustness was demonstrated by generating Z’ value greater than 0.54.
Competition assays have:
- Confirmed the specificity of the detection.
- Demonstrated the advantage of using Alpha by using less binding elements and at least 2-fold less nuclear extract than required to study interactions between transcription factors and their respective DNA sequence with methods commonly used.
Tips
- We offer anti-DIG Alpha beads and streptavidin-coated Alpha beads for association of digoxigenin-labeled oligos and biotinylated oligos, respectively.
- Alternatively, DNA can be directly conjugated to beads using amine-modified DNA oligos and performing the standard reductive amination. This method is more labor-intensive and requires more oligonucleotide, and therefore is not usually recommended.
- For more information on bead conjugation, refer to the Alpha bead conjugation page.
- We recommend including a spacer of around 20 atoms (such as a TEG spacer) between the biotin molecule and the DNA sequence to permit a better accessibility of the DNA strand for its interaction with proteins. For our assay assessments, we used dsDNA containing 20-35 base pairs.
- Result reproducibility may be significantly affected by the use of nuclear extract preparations provided by different suppliers and/or by the use of different nuclear extract lots.
- The specificity of the antibody for its transcription factor has a significant impact on the level of counts generated and therefore, on the assay window. The signal to background ratio (S/B) is calculated as counts obtained in the presence of the consensus DNA sequence over those generated with the mutated DNA sequence.
- The assay buffer (25 mM Hepes, 200 mM NaCl, and 0.1% Tween, pH 7.4) and immunoassay buffer performed similarly in the detection of Sp1 and HNF1 in nuclear extracts. However, the assay buffer may require optimization for the detection of other transcription factors.
- In some cases, it may be worth testing the addition of the pre-mix, composed of the Ab and the AlphaLISA acceptor bead, prior to the addition of oligonucleotides.
- We recommend starting with 20 µg/mL Donor beads and 20 µg/mL Acceptor beads in the reaction. Bead concentration can be titrated later in the assay development (in a cross-titration matrix, titrating each bead from 10 µg/mL to 40 µg/mL).
- The theoretical maximum capacity of a streptavidin-coated Donor bead at 20 µg/mL is 30 nM. Please note that this does not take into consideration the size of the biotinylated molecule that will associate with the bead. For example, depending on the size of the biotinylated molecule, it may saturate the bead at 2-3 nM, rather than at 30 nM. If the Donor bead is saturated, a hook effect may be observed.
- The theoretical maximum capacity of an antibody-coated Acceptor bead is 3-10 nM. Please note that this is a theoretical number —more or less than 10 nM "antigen" may be added to an assay before hooking occurs depending on how strong the antibody-antigen interaction is.
Troubleshooting
View our general Alpha troubleshooting tables.
Citations
View a brief list of citations for Alpha assays.
For research use only. Not for use in diagnostic procedures. The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.




























