Overview
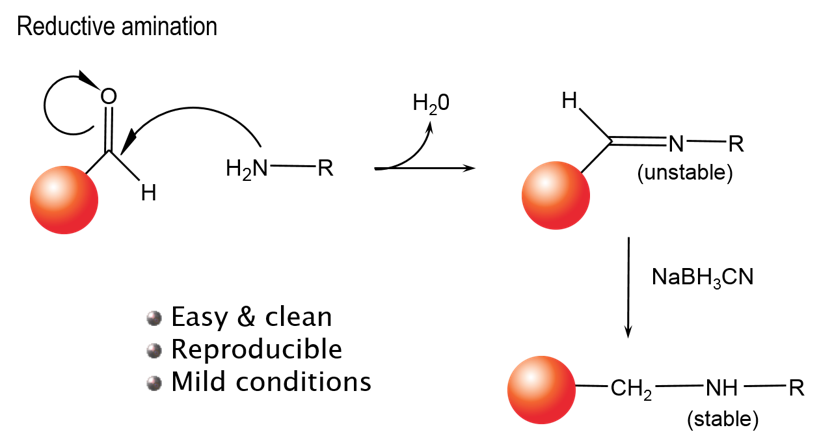
You can use our unconjugated beads to directly conjugate a peptide, protein, or antibody of interest directly to an Alpha Donor, AlphaScreen™ Acceptor, or AlphaLISA™ Acceptor bead. Our unconjugated beads can be used in a reductive amination reaction to link your reagent via free amines (lysines, N-termini) as shown in Figure 1.

Figure 1. Reductive amination reaction for bead conjugation.
What do I need to perform a bead conjugation?
You will need:
- Unconjugated beads (Alpha Donor, AlphaScreen Acceptor, or AlphaLISA Acceptor unconjugated beads)
- Sodium cyanoborohydride powder (Sigma catalog number 156159)
- Carboxymethylamine for blocking (Sigma catalog number C13408)
- Reaction buffer (we recommend 100 mM Hepes pH 7.4)
- Purification buffer (we recommend Tris, pH 8.0)
- Storage buffer (we recommend PBS + 0.05% Proclin® 300)
- Zeba™ desalting columns (Pierce catalog number 89882, 89889, 89891, or 89893 depending on volume)
- Sonicator
- Centrifuge/microfuge
- Vortex
Protocol
- See our ELISA to AlphaLISA immunoassay conversion guide for information, including details for antibody conjugation to beads.
FAQs
Q. What is the bead conjugation reaction?
A. The reaction is a reductive amination. Conjugations will occur on free amine groups, such as lysines.
Q. What will interfere with the conjugation reaction?
A. Glycerol and any free amines (such as Tris, Bis, glycine, etc.) in your sample need to be removed by buffer exchange prior to conjugation. Also, note that your sample should not contain any carrier proteins (BSA, gelatin, etc.), as these would also become conjugated to the beads.
Q: What factors affect the conjugation efficiency?
A: The number of molecules you can conjugate is related to size. The structure will also sterically affect the coupling. Finally, the charge might increase or decrease the proximity, therefore affecting efficiency. Conjugation efficiency is not an easy parameter to measure. You can try to check the conjugation efficiency by decreasing the amount of protein to see if the signal intensity gets reduced.
Q. Do I have to sonicate the beads, as mentioned in the protocol?
A. Bead sonication is not mandatory, yet it is good practice. Centrifugation is the easiest method to wash beads, but centrifuged beads might have some level of clumping. Slightly clumped beads will perform normally, yet might sediment faster due to their “larger” size. We recommend you sonicate after centrifugation. We sonicate using a probe sonicator, by immersing the probe into the bead suspension and sonicating for 20 pulses (counting 1 second per pulse and 1 second between each pulse).
Q. What type of sonicator do you use at Revvity?
A. The probe sonicator used by Revvity manufacturing is a Fisher Scientific, Sonic Dismembrator, Model 500 with a tapered microtip. The Revvity R&D team uses a model that is smaller (hand-held model 60 from the same company).
Q. The technical data sheet for the AlphaScreen Acceptor beads has a slightly different protocol than the one in the AlphaLISA Assay Development Guide for bead conjugation. Does it matter which protocol I follow?
A. You can use either the protocol in the AlphaLISA Assay Development Guide, or the protocol on the AlphaScreen bead technical data sheet for bead conjugation. The protocol in the AlphaLISA Assay Development Guide provides more detail. Some scientists prefer not to do a 48-hour incubation, or wish to conserve antibody.
| AlphaScreen bead coupling protocol | AlphaLISA bead coupling protocol |
|---|---|
| Buffer: 0.1 M MES, pH 6.0 | Buffer: 100 mM Hepes pH 7.4 (this used to be 130 mM sodium phosphate, pH 8.0) |
| Coupling ratio beads:Ab (w/w) at 20:1 | Coupling ratio beads:Ab (w/w) at 50:1 |
| Beads: 5 mg/mL in the labeling mix | Beads: 25 mg/mL in the labeling mix |
| Incubation: 48 hrs at 37°C | Incubation: 18 hrs at 37°C |
Q: Antibodies are normally stored in glycerol. Does glycerol really interfere with the conjugation procedure?
A: At 5% glycerol, we see a small effect on the coupling reaction. At 10%, we see a 2-3x decrease in maximum signal and a 2-3x decrease in sensitivity. You can remove glycerol (when above 5% final) by performing buffer exchange twice using a Zeba™ spin column.
Q: Can I do a thiol-linkage to conjugate to beads?
A: Our standard unconjugated beads have aldehyde reactive group. You need to transform these groups to amine groups to be able to use SIAX (a succinimidyl iodoacetyl amino hexanoate coupling reagent). The amine beads can be generated by reacting the aldehyde beads with ethylenediamine. Also, L-cysteine will be used as the blocking solution, instead of CMO. The method used to conjugate the beads involves transforming a regular AlphaScreen bead into a thiol-binding bead. The protocol involves two steps (amination then activation with sulfo-SIAB) and is very straightforward.
Q: Are sugar stabilizers (e.g. trehalose etc) a problem when conjugating?
A: We have not studied this carefully, but we do not see any theoretical (that’s why it was not studied) or practical (we use trehalose containing antibodies all the time) limitations.
Q: Can we conjugate Fab fragments, rather than whole antibodies?
A: F(ab) fragments conjugated to Acceptor beads should work fine.
For research use only. Not for use in diagnostic procedures. The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.




























