DNA methylation
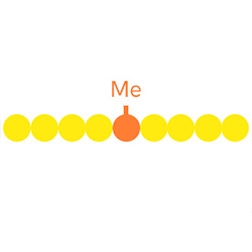
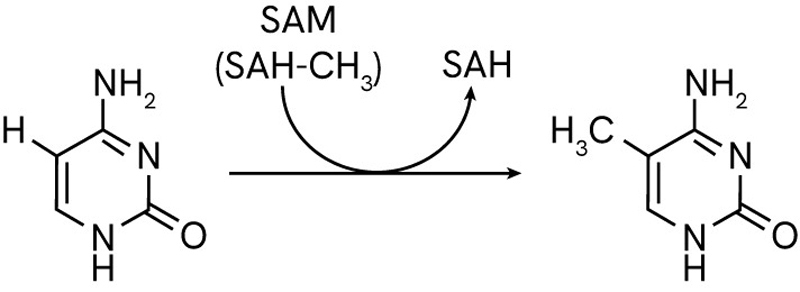
DNA methylation is the main type of epigenetic modification that regulates gene expression and maintains the stability of the genome. It is the process by which a methyl group (CH3) is added to the C5 position of cytosines. This occurs mainly where a cytosine is followed by a guanine (CpG sites) in a DNA sequence, although non-cCpG methylation occurs in plants and sometimes in mammals.
DNA methylation is the main type of epigenetic modification that regulates gene expression and maintains the stability of the genome. It is the process by which a methyl group (CH3) is added to the C5 position of cytosines. This occurs mainly where a cytosine is followed by a guanine (CpG sites) in a DNA sequence, although non-cCpG methylation occurs in plants and sometimes in mammals.
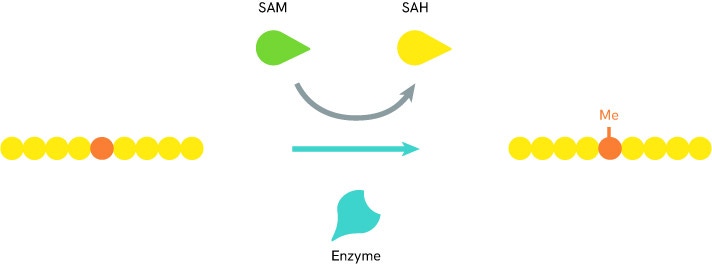
The DNA methylation is performed by a dedicated family of enzymes called DNA methyltransferases (DNMTs). These exist as different types carrying specialized roles with tightly regulated activity and control from internal factors such as the local availability of methylation reaction co-factor S-adenosylmethionine (SAM).
Other proteins are capable of DNA methylation, albeit in a less significant capacity, such as the MBD family (methyl-CpG-binding domain).
DNA methylation promotes local DNA compacting, making it less accessible and readable for transcription and replication purposes, which prevents transcription or replication of the DNA in the region where methylation occurs.
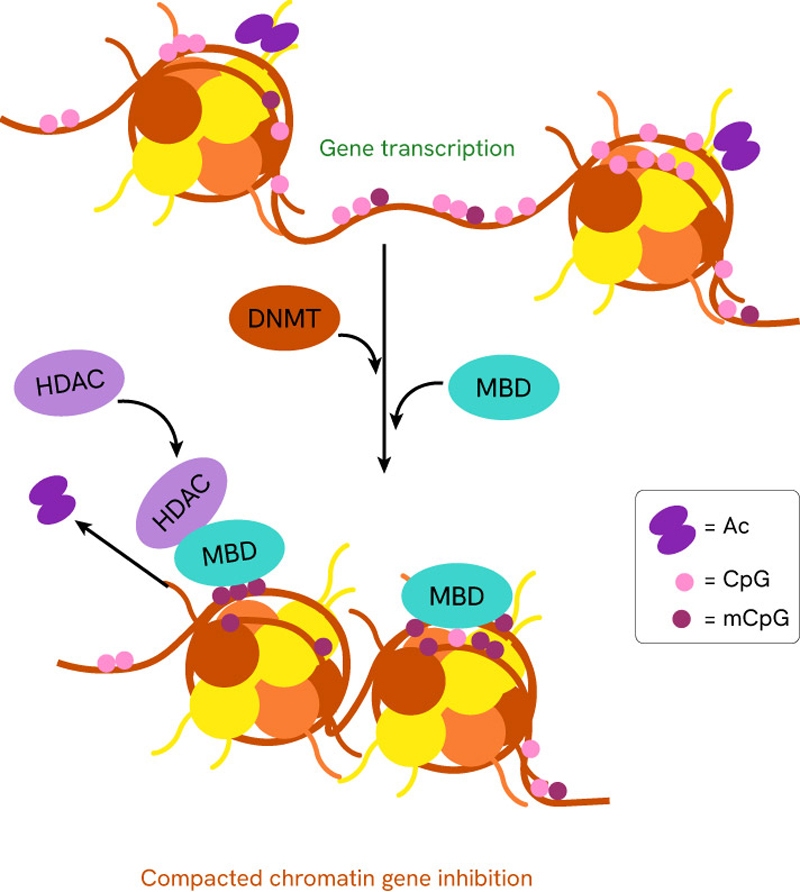
DNA methylation patterns vary by cell type and tissue, allowing cells to specialize in different directions using the same genetic information. Beyond cell types and tissues, DNA methylation is also strongly affected by environmental factors, such as lifestyle, diet, stress, exposure to stress or toxins and UV rays. Changes in DNA methylation have also been linked to biological processes such as embryonic development, aging and certain diseases.
As it is an epigenetic modification, DNA methylation has the potential to transmit from one generation to the next. The differential expression of alleles from the mother or father can even be decided via genomic imprinting. This is where methylated alleles are repressed and genes are expressed differently depending on which parent they are inherited from.
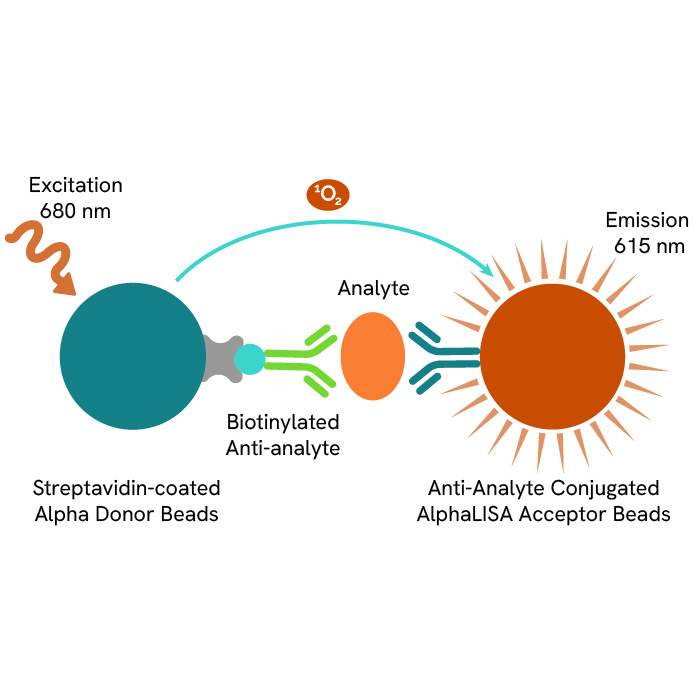
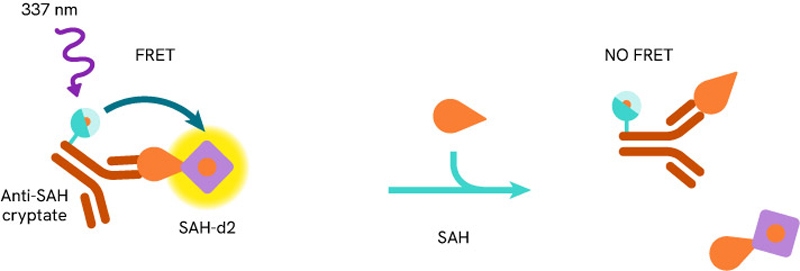
Solutions to detect and measure DNA methylation include methyltransferase assays such as the Revvity’s EPIgeneous™ Methyltransferase Assay. It is a universal biochemical assay for all enzymes within the histone (HMTs) and DNA (DNMTs) methyltransferase families that produce S-adenosylhomocysteine (SAH). Methyltransferase activity is assessed by measuring the conversion of SAM (S-5-adenosyl-L-methionine) to SAH, thanks to an anti-SAH antibody labeled with Tb cryptate and a SAH-d2 tracer. Proximity between these reagents produces a FRET signal, which decreases as the SAH released by the enzymatic reaction competes with the SAH-d2 labeled.

Histone modification
Epigenetic gene regulation also is controlled by changes in histones that make up the nucleosome and by histone modification. Histones are a family of proteins with a key role in the organization and regulation of DNA. They exist in five types (H1, H2A, H2B, H3, and H4) and act as spools that DNA can wrap around in a condensed chromatin state.
Epigenetic gene regulation also is controlled by changes in histones that make up the nucleosome and by histone modification. Histones are a family of proteins with a key role in the organization and regulation of DNA. They exist in five types (H1, H2A, H2B, H3, and H4) and act as spools that DNA can wrap around in a condensed chromatin state.
Histones can be subject to modifications such as methylation, acetylation, phosphorylation, ubiquitination and sumoylation which all affect their ability to assemble, disassemble, and stick to DNA. All of these properties regulate the physical access of proteins to the genome. Because of the way histones condense and wrap DNA around themselves, they oversee which portions of it are accessible to transcription enzymes and other partners that modify or replicate it. By sliding along the DNA sequence, histones cover and uncover different sections prompting the expression of new sequences and the repression of others. For these reasons, histone modifications are critical to gene silencing and expression.
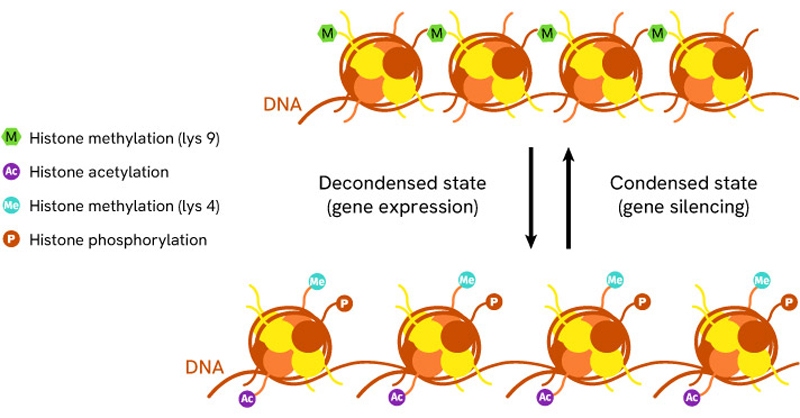
Fig: Histone modifications determine how tightly histones interact with DNA. Transition between different rates of methylation, acetylation and phosphorylation is key in determining the condensation state of DNA and its availability for transcription
This role of histones in DNA regulation is postulated under the “Histone code” hypothesis. H3 and H4 acetylations are the most investigated histone modifications and are found to promote the transcription of genes consistently. Histone methylation on lysine residues is the second most commonly studied histone modification, especially on H3, and is a promoter of DNA condensation that limits DNA access and represses gene expression.
The complexities of histone methylation leave many unknowns about its exact effects. For acetylation and methylation, there are opposite processes of de-acetylation and de-methylation performed by histone deacetylases (HDAC) and demethylases, respectively, which open potential avenues for future therapeutic regulation.
Over the years and using our HTRF and ALPHA technology, Revvity has built a world-class platform of no-wash assays to study epigenetics in cell-based or biochemical formats. Over a dozen of ready-to-use cell-based Histone modification assays address the histone modifications most common and relevant to Epigenetics research, while a wide array of labeled antibodies and reagents constitute a robust toolbox for more custom applications
Noncodings RNA
Expression of noncoding RNAs also plays a role in epigenetic gene regulation. Non-coding RNAs (ncRNAs) are RNA strands that do not code for protein sequences and are not translated into proteins, even though they are transcribed from DNA like all other RNA. Rather, they fulfill an array of functions related to the regulation of cellular processes.
Expression of noncoding RNAs also plays a role in epigenetic gene regulation. Non-coding RNAs (ncRNAs) are RNA strands that do not code for protein sequences and are not translated into proteins, even though they are transcribed from DNA like all other RNA. Rather, they fulfill an array of functions related to the regulation of cellular processes.
With epigenetic mechanisms, ncRNAs are understood to contribute to the regulation of chromatin structure and therefore gene expression via DNA accessibility and readability. They fulfill these roles via direct interaction with other nucleic acids (DNA, RNA) and proteins. There are different types of ncRNAs, characterized by their size and properties:
- Long non-coding RNAs (lncRNAs) are strands that are longer than 200 nucleotides and interact with various complexes that modify chromatin.
- MicroRNAs (miRNAs) are shorter RNA strands of about 22 nucleotides. They play a significant role by binding to specific target messenger RNAs (mRNAs) and downregulating their stability and/or translation rate.
- Small interfering RNAs (siRNAs) are similar in size to miRNAs and share similar functions in the context of epigenetics as they promote gene silencing by targeting specific mRNA for degradation or translational repression.
The precise mechanisms by which ncRNAs regulate epigenetic processes remain in parts undescribed. With cell-type and tissue-dependent functions, they are reliant on environmental factors and developmental stages. Two main axes seem to be involved in the recruitment and modeling of regulatory complexes by lncRNAs, and the promotion/inhibition of histone modification and DNA methylation by miRNAs and siRNAs.


Filters
1 - 25 of 124 Products and Services
The HTRF SIRT1 assay enables a straigthforward screening and characterization of compounds activating or inhibiting SIRT1 activity
The Total H3 cellular assay is optimized for the detection of endogeneous levels of Histone H3 in cells.
This kit is designed for the detection of di-methylated Histone H3 Lysine 4 (H3K4me2) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
This kit is designed for the detection of acetylated Histone H3 Lysine 9 (H3K9ac) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
This kit is designed for the detection of di-methylated Histone H3 Lysine 36 (H3K36me2) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
This kit is designed for the detection of tri-methylated Histone H3 Lysine 27 (H3K27me3) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
This kit is designed for the detection of di-methylated Histone H3 Lysine 9 (H3K9me2) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
This kit is designed to be used as a control for the AlphaLISA Epigenetic Cellular Detection Kits, through detection of unmodified Histone H3 Lysine 4 (H3K4) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
AlphaLISA™ Acceptor beads conjugated to an antibody against human histone H3 di-methylated at lysine 9 (H3K9me2). These beads can be used for no-wash AlphaLISA epigenetic writer and eraser assays.
Terbium-labeled streptavidin for binding biotinylated biomolecules in DELFIA® time-resolved fluorescence (TRF) assays.
Europium-labeled monoclonal antibody recognizing human Histone H3 di-methylated at lysine 36 (H3K36me2). For use in LANCE® Ultra epigenetic writer and eraser assays.
Europium-labeled monoclonal antibody recognizing human Histone H3 and H4 methylated arginine residues. For use in LANCE® Ultra epigenetic writer and eraser assays.
Europium-labeled monoclonal antibody recognizing human H3 mono- and di-methylated at lysine 4 (H3K4me1-2). For use in LANCE® Ultra epigenetic writer and eraser assays.
This Eu-cryptate conjugate allows specific detection of the K9Me1 motif on histone H3, as a result of methylation or demethylation enzymatic activity.
Europium-labeled monoclonal antibody recognizing human Histone H4 (H4Kac pan). For use in LANCE® Ultra epigenetic writer and eraser assays.
This cell-based assay format enables the detection of the H3K27Me3 motif, as a result of histone H3 methylation.
The EPIgeneous methyltransferase assay is a kit optimized for the assessment of enzymatic methylation activity through S-adenosylhomocysteine detection (SAH).
This biochemical assay format enables the detection of the interaction between a subset of epigenetic binding domains and modified histone sequences.
This biochemical assay format enables the detection of the interaction between a subset of epigenetic binding domains and modified histone sequences.
This cell-based assay format enables the detection of the H3K36Me2 motif, as a result of histone H3 methylation by EZH2 enzyme among others.
This kit is designed for the detection of acetylated Histone H3 Lysine 27 (H3K27ac) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
This biochemical assay format enables the detection of the interaction between a subset of epigenetic binding domains and modified histone sequences.
This cell-based assay format enables the detection of the H3K4Me2 motif, as a result of histone H3 modifications by SET or LSD1/2 enzymes among others.
This kit is designed for the detection of di-methylated Histone H3 Lysine 79 (H3K79me2) in cell lysates using a homogeneous AlphaLISA assay (no wash steps).
AlphaLISA no-wash assay kit for the detection of total human Histone 2A (H2A) in cell extracts or cells in culture medium.