Gene functions can be determined in a dynamic living system through systematic knockdown, knockout, or overexpression of genes. This experimental approach is known as functional genomics and has become a key discovery tool in many areas of biological research, such as drug target identification, drug resistance, host‑pathogen interactions, and biological pathway analysis.
In 2005, Dharmacon, Inc. - now part of Revvity - produced the first siRNA reagent library to target each gene in the human genome. Since then, whole-genome libraries have enabled individual down-regulation of thousands of genes in parallel, providing a revolutionary strategy for functional genomics studies beyond gene expression microarrays.
Today, high-throughput genetic screening technologies include libraries of small interfering RNA (siRNA), short hairpin RNA (shRNA), microRNA, or CRISPR-Cas9 guide RNA (gRNA). By integrating sophisticated automation, biological assays, high-content data capture, and robust bio-statistical analysis, these approaches are routinely employed in academic, pharmaceutical, and biotechnology laboratories, becoming a cornerstone in modern life sciences and drug discovery.

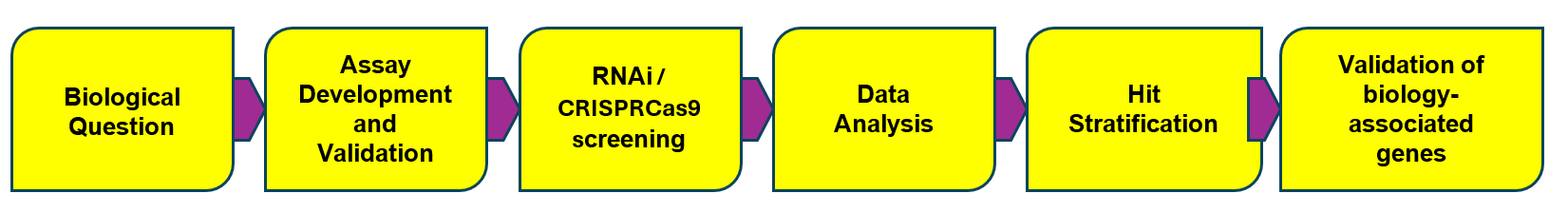
General Screening Workflow. The success of RNAi and CRISPR-Cas9 screening hinges on meticulous testing, careful planning, and thorough implementation at every critical phase. Each step is crucial to ensure reliable and meaningful results.
Discussion has emerged about the advantages of modern CRISPR over the siRNA technologies for functional genomic screening1. The siRNA is considered easier to use, with straightforward synthesis and delivery into cells using standard transfection methods. Because of the transient nature of the gene silencing, siRNA can help study short-term loss of function. This can also be considered a disadvantage, where the effects of siRNA can typically last only for a few days, which cannot be suitable for long-term studies2. Also, siRNA libraries are less expensive compared to CRISPR libraries.
CRISPR-Cas9 offers several advantages, including creating permanent gene knockouts, which allow for long-term gene function studies. It can be designed to target specific genomic loci with high precision. It is versatile and is used for various applications such as gene knockout, gene activation (CRISPRa), and gene interference (CRISPRi).
However, there are also disadvantages to consider. Despite its high specificity, CRISPR-Cas9 can still cause off-target mutations, leading to unintended consequences. Efficient delivery of the CRISPR-Cas9 components into cells can be challenging, especially in specific cell types. The potential for germline editing and heritable genetic modifications raises ethical concerns. Both siRNA and CRISPR-Cas9 have their unique strengths and limitations, and the choice between them often depends on the specific requirements of the study.
Both siRNA and CRISPR-Cas9 have unique strengths and limitations, and the choice between them often depends on the specific requirements of the study3.
Choosing the right reagent libraries for functional genomics
The choice of screening library depends heavily on the goals of the intended experiments. Scientists must carefully consider what they aim to achieve to determine which library type will be most suitable. When it comes to targeting mammalian genomes for gene knockdown or knockout, scientists have a variety of reagent libraries at their disposal. These include siRNA, shRNA, microRNA, and CRISPR-Cas9 sgRNA libraries, available from Revvity.
Other important factors include the experimental goals, reagent design, production quality control, and the level of scientific support provided by the library source. Each library type offers unique advantages in specific contexts, making it essential to choose wisely.
Combining libraries for optimal results
Many advanced functional genomics screening laboratories use a combination of siRNA, shRNA, microRNA, and CRISPR-Cas9 gRNA libraries. This approach allows researchers to leverage the strengths of each type, ensuring comprehensive and robust results2. By thoughtfully selecting and combining these powerful tools, scientists can push the boundaries of biological research, uncovering new insights and driving innovation in the field.
Effective delivery and timing of gene silencing
In most cell types, delivering siRNAs is straightforward with the help of transfection reagents. Typically, these siRNAs just need to be resuspended in water or buffer before use. For cells that are difficult to transfect, some siRNAs have been chemically modified to bypass the need for lipid transfection reagents or electroporation.
Gene silencing usually peaks within 48-72 hours and fades away by 5-7 days in actively dividing cells. However, this knockdown duration might not always be sufficient depending on the specific assay or phenotype being measured.
By understanding these nuances, researchers can better plan their experiments and choose the right tools to achieve their scientific goals.
Plasmid libraries
shRNA libraries can be supplied as plasmid-transformed E. coli glycerol stocks microtiter plates. This setup provides a renewable source of RNAi material but has some challenges. Using these libraries is labor-intensive. Each clone must be grown and maintained to ensure quality and viability. Additionally, preparing the plasmids for use is a time-consuming process. The gene silencing achieved with shRNA is transient and depends on the shRNA's cellular expression level. Therefore, selecting a promoter with high activity in the cell type under study is crucial for effective results. By understanding these aspects, researchers can better navigate the complexities of using plasmid libraries and optimize their experiments for successful outcomes.
Lentiviral libraries
Viral delivery methods are recommended to harness the full potential of shRNA. These methods allow for efficient transduction of many cell types, including primary and non-dividing cells, using shRNA vector constructs packaged into non-replicating lentiviral particles. However, producing lentiviral particles in 96-well plates can be both challenging and expensive.
For arrayed screening, the workflow requires a variety of instruments to handle liquid, stack plates, manage lids, and provide sufficient incubator capacity for numerous plates. Assay read-out equipment and robust data management systems are also essential. Robotic systems are often employed to boost accuracy and reproducibility, especially in large-scale screens like whole-genome or druggable target screens. Techniques such as reverse transfection help minimize liquid handling steps, reducing variability.
It can be technically tricky to control variations between wells and plates. Achieving high titers means individually packaging each construct and arranging the viruses into a proper screening format. Because of these hurdles, while lentiviral shRNA approaches are well-suited for observing long-term, integrated silencing effects, the technical limitations can make this high-throughput genome-wide screening strategy costly and labor-intensive.
Pooled lentiviral libraries
Pooled lentiviral shRNA methods combine the benefits of lentiviral delivery with high-throughput screening. This strategy involves mixing hundreds or thousands of unique shRNAs or sgRNAs into one pool and then packaging them into lentiviral particles. This way, cells can be transduced in larger groups, selection pressure can be applied, and cells expressing individual hairpins can be isolated and identified using various selection strategies. This innovative approach doesn't require liquid-handling robotics, making it accessible to more research groups. Pooled lentiviral shRNA screening has already led to numerous published functional genomic screens, both in vitro and in vivo.
By leveraging these advanced techniques, researchers can delve deeper into the complexities of gene function and regulation, paving the way for groundbreaking discoveries in functional genomics.
Perspectives for functional genomic screening
High-throughput functional genomics screening has become a staple in understanding molecular mechanisms in both normal and disease states. This powerful technique involves systematically knocking down or overexpressing genes using siRNA, shRNA, microRNA, and CRISPR-Cas9 guide RNA libraries. The goal? To observe measurable phenotypes that reveal the roles of these genes.
Functional genomic screens demand more rigorous assay validation and optimization when it comes to collecting phenotypic data compared to single-gene experiments. Standard lab equipment like plate readers can be used for phenotypic read-outs from RNAi and CRISPR-Cas9 screens. These read-outs often involve colorimetric, fluorescent, or luminescent assays to measure cell proliferation, protein secretion, reporter gene activity, and apoptosis.
Recently, high-throughput functional screening has advanced with the inclusion of automated fluorescent microscopy platforms and sophisticated high-content imaging tools. These technologies capture spatial, temporal, and kinetic multi-parametric data sets, enhancing the sensitivity of screens by allowing detailed characterization of loss-of-function phenotypes in a broader biological context.
With the advent of machine learning technologies, functional genomic editing will pave the way for comprehensive modeling by artificial intelligence models, which can use the genome as the blueprint for large language models4.
Get help with your functional genomics project from our preclinical services experts today.
References:
- Haley & Roudnicky. 2020. Functional Genomics for Cancer Drug Target Discovery. Cancer Cell.
- Campeau & Gobeil. 2011. RNA interference in mammals: behind the screen. Brief. Funct. Genom.
- Taylor & Woodcock. A Perspective on the Future of High-Throughput RNAi Screening: Will CRISPR Cut Out the Competition or Can RNAi Help Guide the Way? J Biomol Screen. 2015
- Sarumi & Heider. 2024. Large language models and their applications in bioinformatics. Comput. Struct. Biotechnol. J.

































