Clinical researchers and their affiliated laboratories are crucial to accelerating potential medical discoveries that ultimately benefit society and the well-being of its people. Research is active in generating meaningful insights into disease processes for new drug design and biomarker detection that could potentially help improve data-driven decisions in the stratification strategies for complex disorders. Within precision medicine research, having an informed prediction of response and tolerance to a treatment plan is critical as new therapies progress into potential clinical practice. In fact, 25% of all new FDA-approved drugs in 2019 were personalized medicines; in the same vein, 65% of approval requests to the EMA and FDA between 2015 - 2019 included at least one biomarker consideration within the drug development process1. Thus, the momentum behind in vitro diagnostic (IVD) research – and how to go about its adoption and implementation – shows no sign of slowing.
Innovation extends into a less visible but just as important prong of operational and procedural due diligence for laboratory scientists. The implications of what it means to adopt a sustainable and scalable lab process can be wide and far-reaching, especially when we scale into realms of applications benefiting public health and safety. In fact, the world needs no reminder of the value that laboratory readiness can bring when it comes to infectious disease outbreaks at a time where we are experiencing global changes in climate, technological development / globalization, and population composition / urbanization2.
In short, the rigor of the scientific method is used as well, when communities, institutes, and labs together plan on researching smart ways to scale workflows. There are technical considerations lab management typically builds into their plans ahead of time:
- Sample type – access, availability, transportability, and storage. Especially important for repeat sampling requirements, cross-site technical validation of assay performance, re-calibrations, etc.
- Sample prep – ease-of-processing raw sample to analyte, maximum time permitted between collection and analyte extraction, quantity and quality required as assay input to generate useful data, etc.
- Sample throughput – ease of scaling, ease of migrating complete processes or specific bottlenecks to automation, expectations in start-to-finish turn-around time, contingency for sample diversion, etc.
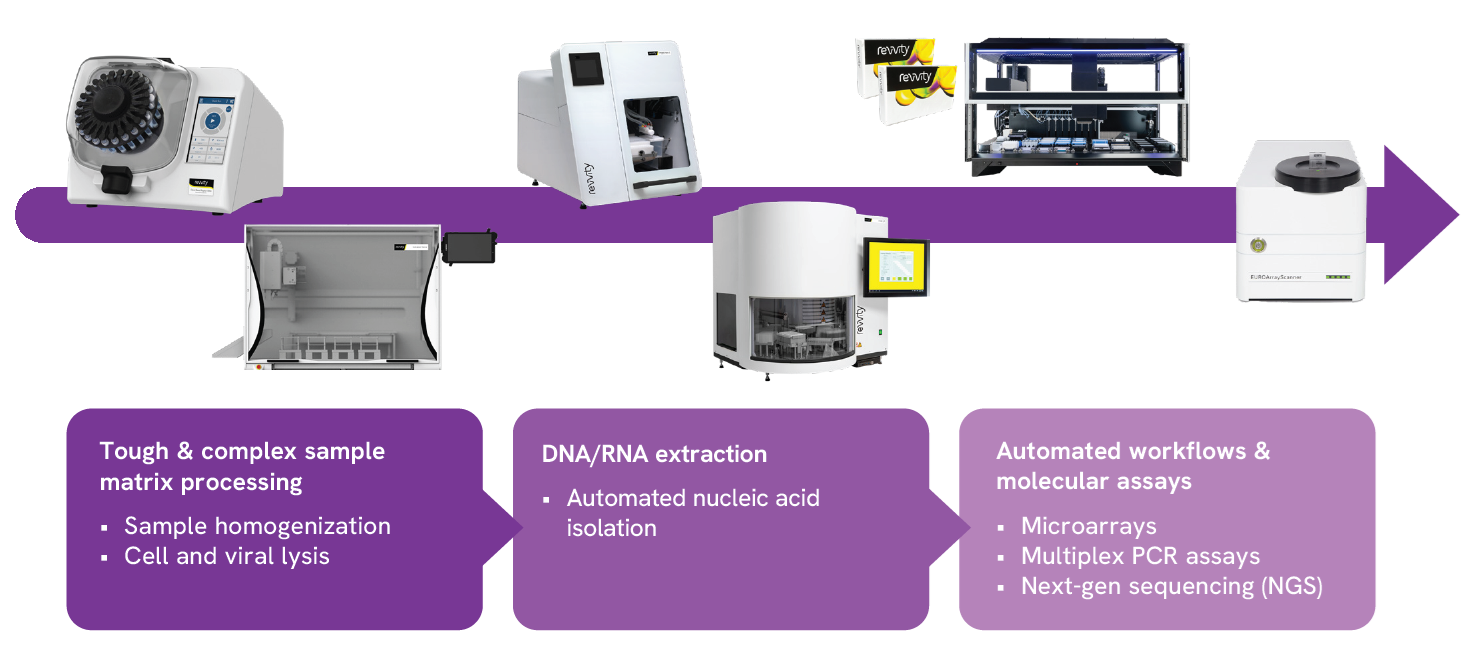
With each, there are accompanying metrics that are measurable to help gauge the health and efficiency of a lab operation and to make calculated, data-driven decisions when changing or adopting a new workflow. Any good measure will also help assess risk – risk to sample, risk to result, risk attributed to human error, and so forth. Another important benchmark most labs keep on their kanban or in their daily standing meeting is the number of samples processed, as well as improvements to average turn-around time (TAT). When maintaining a lean crew, tools that offer robotic-repeatability and time-savings can make a big difference to the bench, especially when working with a variety of complex tissues or tough matrices.

Read this application note showing how bead mill homogenization reduces sample prep from fingernails down to 80 mins, compared to overnight incubation of 13-16 hours, for dermatomycoses organism identification via a PCR-based microarray analysis.
Research use only. Not for use in diagnostic procedures.
References:
- Valla, V., Alzabin, S., Koukoura, A., Lewis, A., Nielsen, A. A., & Vassiliadis, E. (2021). Companion Diagnostics: State of the Art and New Regulations. Biomarker insights, 16, 11772719211047763. https://doi.org/10.1177/11772719211047763
- Baker, R.E., Mahmud, A.S., Miller, I.F. et al. (2022). Infectious disease in an era of global change. Nat Rev Microbiol 20, 193–205. https://doi.org/10.1038/s41579-021-00639-z

































