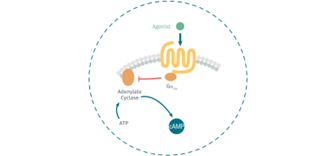
What is the role of Gαi/o coupled GPCRs?
Gαi/o protein is implicated in the regulation of cAMP production:
- In presence of an agonist of GPCRs, it inhibits adenylate cyclase (AD).
- In presence of an antagonist of GPCRs, it cannot inhibit AD.
So, the addition of an agonist to your assay will mean a decrease in the cAMP concentration. On the other hand, if you add an antagonist you will observe an increase in the cAMP concentration in your plates.

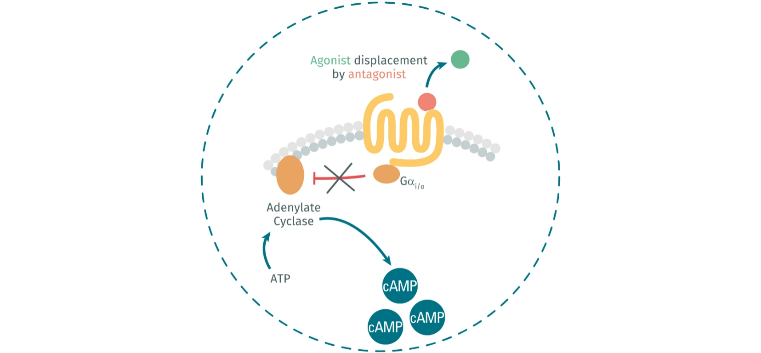
Gαi/o protein role in the presence of an agonist and antagonist
When it comes to optimizing a cAMP Gi assay, there are several parameters to consider. One of the first steps is to take good care of your cells, and to optimize their preparation.
This step completed; other elements need to be considered.
It’s time to focus on the forskolin concentration!
What is Forskolin (FSK)?
Forskolin is known to be a full agonist of AD.
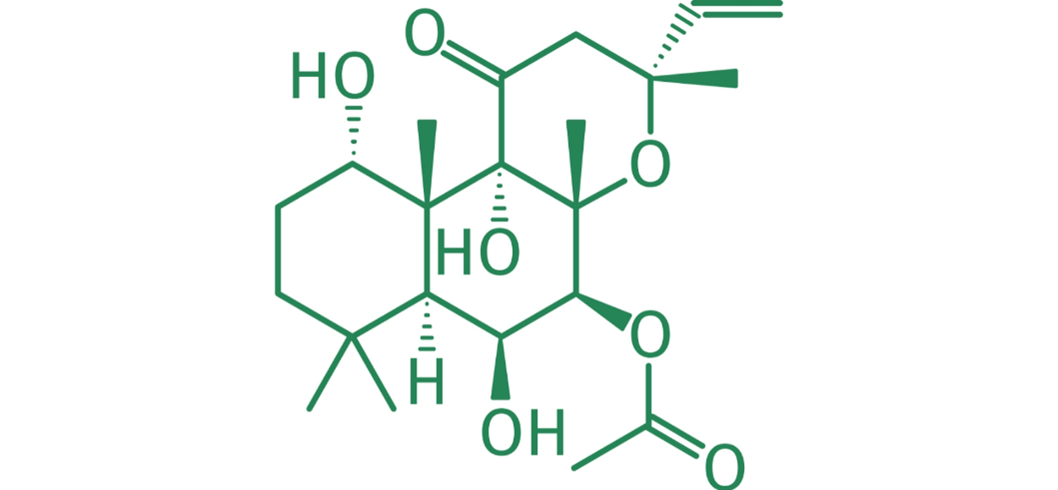
Representation of the forskolin molecule
It is widely used for Gi coupled receptor studies in a pre-activation step to reveal a cAMP inhibition upon cell stimulation. Optimization of FSK is a way to minimize errors and obtain better results. During the optimization, we are looking for the maximal assay window.
The method consists in doing experiments for each cellular density, assaying a full dose response of the forskolin alone (basal) or pretreated with a maximal dose of the agonist (treated cells).
To give you an example and show you the importance of this titration, our experts did these experiments for 4 cellular densities from 2 000 to 8 000 cells/well, and 9 forskolin concentrations from 0 to 500 μM.
| Assay window(Ratio treated cells/Ratio basal) | ||||
|---|---|---|---|---|
| [FSK](μM) | 2 kcells/w | 4 kcells/w | 6 kcells/w | 8 kcells/w |
| 0 | 1 | 1 | 1 | 1 |
| 0.025 | 1 | 1 | 1 | 1.1 |
| 0.25 | 1.1 | 1.3 | 1.5 | 19 |
| 0.51 | 1.3 | 1.6 | 2 | 2.7 |
| 2.5 | 2.2 | 3.2 | 4.2 | 4.5 |
| 5 | 2.5 | 3.6 | 4.3 | 4.4 |
| 12 | 2.5 | 2.8 | 2.9 | 2.7 |
| 25 | 1.7 | 1.5 | 1.4 | 1.4 |
| 500 | 1.9 | 1 | 1 | 0.9 |
Table: Experiment results depending on cell densities and forskolin concentrations
For these experiments, the best compromise is 6 000 cells/well treated with a dose of forskolin of between 2.5 and 5 μM. If you took the highest cell concentration illustrated here, the risk would be to be too close to what is called the hook effect, so it’s better to be just before the plateau and the arrival of the hook effect.
Optimize your stimulation buffer, cell densities, and stimulation time
The optimization of these three parameters will help you minimize errors when you want to quantify cAMP accumulation in response to Gαi/o coupled GPCR activation or inhibition. We have developed optimization content for the study of Gαs coupled GPCRs which is also useful for Gαi/o coupled GPCRs assays.
Measure cAMP modulations in response to Gi-activation
Based on HTRF™, a reliable technology, the cAMP Gi assay contains all the reagents needed to quantify cAMP, ensuring that you benefit from a complete tool for your needs. This kit is based on a competition between native cAMP produced by cells and cAMP labeled with the cryptate for binding to a d2 labeled antibody.
For research use only. Not for use in diagnostic procedures
The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.