Overview
Bead selection is an important aspect of assay design. Bead selection can impact assay performance, and in some cases, can mean the difference between a successful assay and an unworkable assay. Bead selection is an important aspect of assay design. Bead selection can impact assay performance, and in some cases, can mean the difference between a successful assay and an unworkable assay. An understanding of the various ways bead choice can affect your assay may be helpful when you are planning your experiments. The information that follows is intended to be used to help guide bead selection for your particular assay, though keep in mind there is no substitute for empirically testing the impact of different beads on an assay.
AlphaLISA™ vs. AlphaScreen™ beads
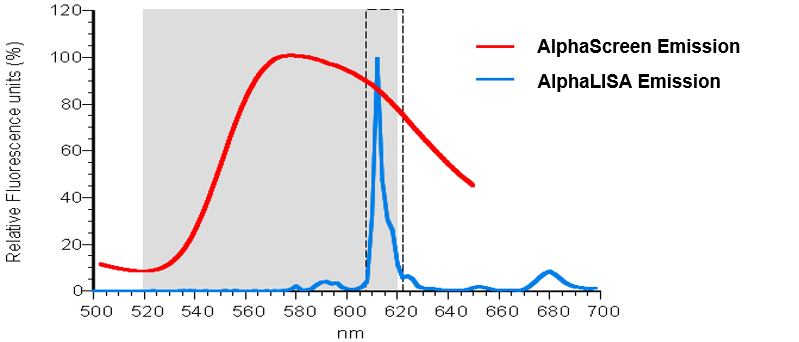
Both AlphaLISA and AlphaScreen technologies rely on the use of the same type of Donor bead: the Alpha Donor bead. However, this Donor bead can be paired with either an AlphaLISA Acceptor bead or an AlphaScreen Acceptor bead. The main difference between these two Acceptor beads is the final fluorophore used to generate signal. AlphaScreen Acceptor beads use rubrene as the final fluorophore, emitting light between 520 and 620 nm. AlphaLISA Acceptor beads use a Europium chelate as the final fluorophore, emitting light in a narrower peak at 615 nm. This makes the AlphaLISA Acceptor bead less prone to interference from buffer components or other chemicals that absorb light between 520 and 600 nm.

Figure 1. Emission spectra of AlphaScreen and AlphaLISA Acceptor beads.
The decision of whether to use an AlphaLISA bead or AlphaScreen bead will likely depend on:
- Sample type. Complex samples (such as serum or plasma) may have components that interfere by absorbing light in the 520-600 nm range. For example, hemoglobin has a broad and intense absorbance spectrum up to 600 nm. If your sample has hemoglobin present, you will want to select AlphaLISA Acceptor beads (rather than AlphaScreen Acceptor beads) to avoid signal interference.
- End goal/purpose. If you are screening libraries of compounds, you may want to select AlphaLISA Acceptor beads for your assay in order to reduce or avoid interference from compounds in the screening library itself. If you were to use AlphaScreen Acceptor beads and a particular compound in your library can absorb light between 520 and 600 nm, you may see reduced signal leading to a false positive or false negative result (depending on how your assay is set up).
- Convenience and type of bead coating offered.
- AlphaLISA beads are offered as individual toolbox reagents (Donor beads are offered separately from Acceptor beads), allowing the ability to mix-and-match bead options.
- AlphaScreen beads are typically packaged as kits, pairing streptavidin Donor beads with AlphaScreen Acceptor beads that bind to a particular tag or label, as indicated in the product name. The AlphaScreen kits also include an appropriate biotinylated “probe” (such as biotinylated-6X His, biotinylated GST tag, etc.) that can be used as a positive control to check that the beads are working correctly.
Bead affinities and binding capacity
Each type of Alpha bead has a characteristic theoretical bead capacity (see Table 1, below). This is the point at which the beads are saturated with associating protein, and additional protein will not be able to associate with the bead. If you have saturated a bead, you may see a “hooking effect”. Below the hook point, both Donor and Acceptor beads become progressively saturated by the target molecule and the signal increases with increasing target concentration. At the hook point, either the Donor or the Acceptor component is saturated with the target molecule and a maximum signal is detected. Above the hook point, an excess of target molecules oversaturates the Donor or the Acceptor beads, which inhibits their association and causes a progressive signal decrease. The hook effect is a common phenomenon found when using any sandwich-type assay (for example, ELISA assays).
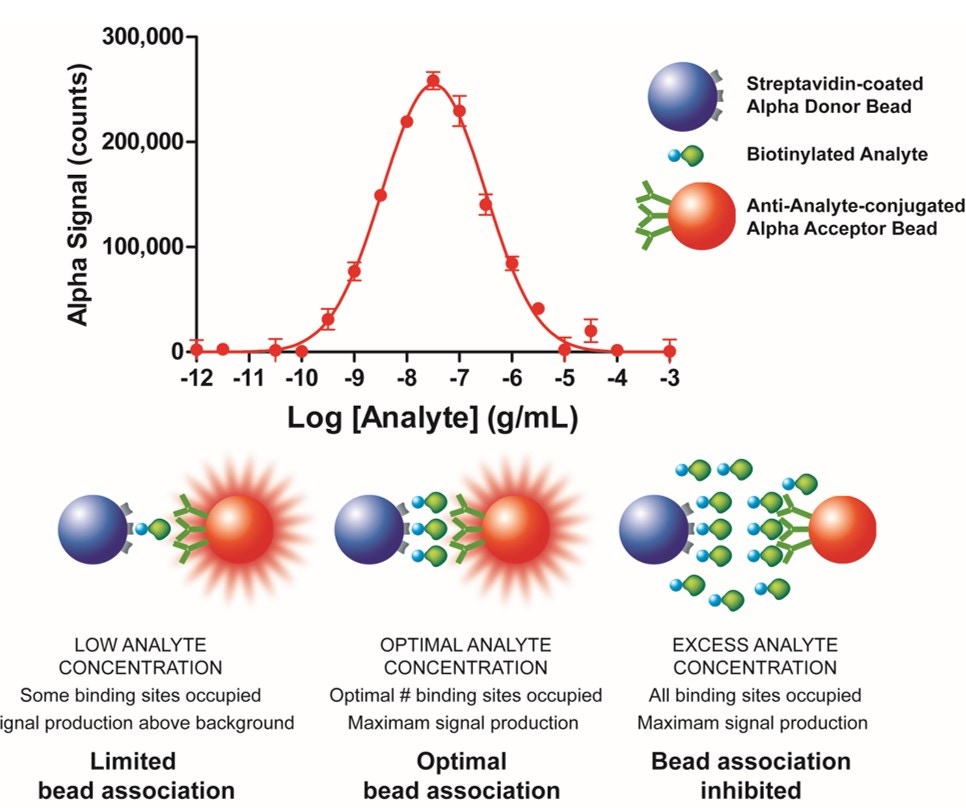
Figure 2. Illustration of the hook effect in a protein detection assay.
Bead capacities are influenced by:
- Size of the biomolecule associated with the bead: more small peptides will fit on the surface of a bead, compared to large antibodies. For example, a streptavidin-coated bead used at 20 μg/mL usually saturates at around 30 nM biotinylated peptide but will saturate at around 2-3 nM biotinylated antibody.
- Affinity of the bead for the associating reagent: for a GST-tagged protein, one can choose either anti-GST antibody-coated beads, or glutathione beads. However, the affinity of glutathione for GST is weaker compared to the affinity of anti-GST antibody for GST. This means that one would be able to add higher concentrations of GST-tagged protein to a glutathione bead before saturating/hooking, because for a given protein concentration more GST-tagged protein is not associated with the glutathione bead. You may find that the hook point is reached on anti-GST beads at around 20 nM GST-tagged protein, but at around 200 nM GST-tagged protein on glutathione beads.
The use of weaker affinity beads can be particularly helpful if you are studying a weak biomolecular interaction (Kd > 100 nM, such as most protein-protein interactions), as you can add more protein to drive the equilibrium to more bi-molecular complex formation before saturating the bead.
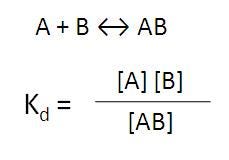
Equilibrium
For strong biomolecular interactions (Kd < 20 nM, such as most receptor-ligand interactions), the protein concentration may not need to be titrated to as high a level, and a higher signal may be obtained with a stronger affinity bead because less protein is dissociated from the bead (more of the interacting biomolecules in the assay are contributing to signal).
Table 1. Relative Capacities of Various Beads (Beads Used at 20 μg/mL Final Concentration in Assay).
|
Bead Coating |
Used to Bind/Capture |
Molecule and Partner Bead Used to Determine Theoretical Binding Capacity |
Theoretical Binding Capacity1(Provided for Relative Comparison) |
|---|---|---|---|
|
Streptavidin |
Biotinylated peptides, proteins, oligos, sugars, small molecules, etc. |
Biotinylated peptide |
30 nM |
|
Strep-Tactin® |
Strep-Tag® II and One-STrEP tagged proteins/peptides |
One STrEP-azurin-6X His with nickel chelate beads (100 nM) |
|
|
Anti-GST antibody |
GST-fusion proteins and peptides |
Biotinylated GST with streptavidin bead |
3 nM |
|
Anti-6X His antibody |
His-tagged proteins and peptides |
6X His-GST with glutathione bead |
100 nM |
|
Anti-FLAG antibody |
FLAG-tagged proteins and peptides |
Biotinylated FLAG with streptavidin bead |
100 nM |
|
Anti-maltose binding protein (MBP) antibody |
MBP-tagged proteins |
Biotinylated MBP with streptavidin bead |
1 nM |
|
Anti-HA antibody |
Hemagluttinin-tagged proteins and peptides |
Biotinylated-PEG-HA with streptavidin bead |
10 nM |
|
Anti-c-myc antibody |
c-myc-tagged proteins and peptides |
Biotinylated c-myc with streptavidin bead |
100 nM |
|
Anti-DIG antibody |
Digoxigenin labeled proteins, peptides oligos, etc. |
Biotinylated digoxigenin with streptavidin bead |
1 nM |
|
Anti-FITC antibody |
FITC or fluorescein-labeled proteins, peptides, oligos, sugars, small molecules, etc. |
Biotinylated-ERE-FITC with streptavidin bead |
>1 nM |
|
Anti-V5 antibody |
V5-tagged proteins/targets |
Biotin-Chromalink V5 (14 aa) with streptavidin bead |
~3 nM |
|
Anti-GFP antibody |
GFP-tagged (green fluorescent protein-tagged) proteins and peptides |
Biotinylated GFP with streptavidin beads |
3 nM |
|
Glutathione (GSH) |
GST-fusion proteins and peptides |
6X His-tagged GST with nickel chelate bead |
300 nM -1 μM |
|
Nickel chelate (Ni2+) |
His-tagged proteins and peptides |
6X His-tagged GST with glutathione bead |
300 nM -1 μM |
|
Protein A |
Antibodies2 |
Biotinylated rabbit IgG with streptavidin bead |
3 nM (antibody3) |
|
Protein G |
Antibodies4 |
Biotinylated rabbit IgG with streptavidin bead |
1 nM (antibody3) |
|
Protein L |
Antibodies5 |
Biotinylated human IgG (kappa) with streptavidin bead |
1 nM (antibody3) |
|
Anti-human IgG |
Fc portion of human IgG antibodies |
Biotinylated human IgG with streptavidin bead |
3 nM (antibody3) |
|
Anti-rabbit IgG |
Fc portion of rabbit IgG antibodies |
Biotinylated rabbit IgG with streptavidin bead |
1 nM (antibody3) |
|
Anti-mouse IgG |
Fc portion of mouse IgG antibodies |
Biotinylated mouse IgG with streptavidin bead |
3 nM (antibody3) |
|
Anti-mouse IgM |
Mouse IgM immunoglobulins |
Biotin-mouse IgM with streptavidin bead |
0.3 nM (antibody3) |
|
Anti-rat IgG |
Fc portion of rat IgG antibodies |
Biotinylated rat IgG with streptavidin bead |
1 nM (antibody3) |
|
Anti-sheep IgG |
Fc portion of sheep IgG antibodies |
Biotinylated sheep IgG with streptavidin beads |
1 nM (antibody3) |
|
Anti-goat IgG |
Fc portion of goat IgG antibodies |
Biotinylated goat IgG with streptavidin bead |
3 nM (antibody3) |
|
Anti-chicken IgY |
Fc fragment of chicken IgY immunoglobulins |
Biotinylated chicken IgY with streptavidin beads |
0.3 nM (antibody3) |
- The numbers provided are examples given to compare relative bead capacities, and are derived from probe titration experiments, as shown in the QC data on the product tech data sheets. The actual bead capacity for your bead system should be determined empirically for a given assay. The bead capacities given in this table are influenced by partner bead and the size of the probe used in these assays. The actual bead capacity will be influenced by size of the protein associated with the bead, as well as affinity of the protein (or antibody) for the bead.
- Protein A interacts strongly with particular subclasses of antibodies, including human IgG1, IgG2, and IgG4; mouse IgG2A and IgG2B; and rabbit, human and mouse total IgG. Protein A also has weaker affinity for other antibody subclasses. The affinity depends on the isotype and species of the antibody associating with the bead. Pierce has a nice chart that may be helpful.
- The bead capacity for these products refers to the capacity for antibody. The amount of protein you will then be able to bind to the antibody on the bead will be dependent on the affinity of your antibody for your protein and the size of your protein, and may range anywhere from 1 pM to 1 μM.
- 4Protein G binds to all subclasses of human IgG and mouse IgG. In addition, it binds to rat, goat, sheep, guinea pig, rabbit, cow, pig and horse antibodies. Pierce has a nice chart that may be helpful.
- Protein L binds efficiently total human IgG, IgM, IgA, IgE, IgD, mouse IgG and rat IgG. It binds only poorly mouse IgM and rabbit IgM and it does not bind human IgG (lambda light chain), rabbit, sheep, goat and bovine IgG and rat IgM. Pierce has a nice chart that may be helpful.
Specific bead pairs that can lead to false-positive signals
In some cases, a particular pairing of Donor and Acceptor beads can cause problems when the antibodies or proteins coated on the two beads have affinity for one another. For example, you could try to develop a sandwiching immunoassay using Protein A-coated Donor beads to capture a rabbit IgG antibody, and anti-human IgG antibody-coated Acceptor beads to capture a human IgG antibody. However, this would be poor assay design, because the Protein A-coated Donor bead can directly interact with the anti-human antibody-coated Acceptor bead, leading to a false positive signal. The selected beads should not be able to directly interact with each other.
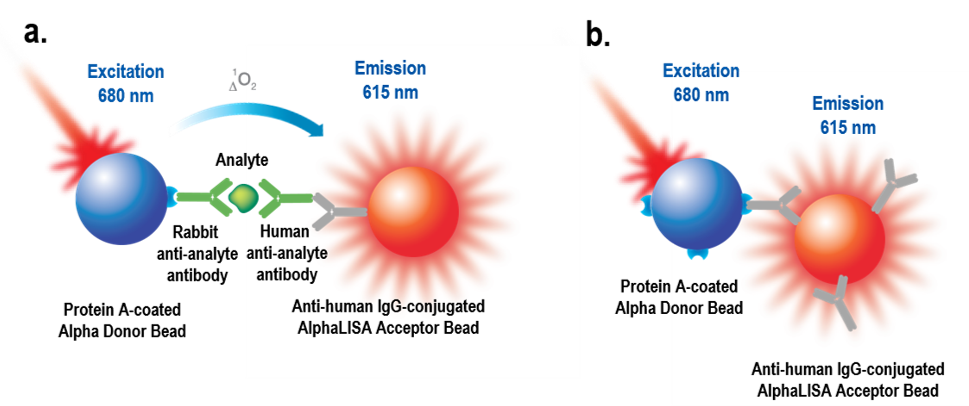
Figure 3: a) Intended assay design for sandwiching immunoassay. Protein A-coated Donor beads are intended to be used to capture a rabbit IgG antibody against an analyte of interest. Anti-human IgG antibody-coated Acceptor beads are intended to be used to capture a human IgG antibody against the analyte of interest to complete the sandwiching assay. b) What can happen given the bead pair choice. The Protein A-coated bead can also bind directly to the anti-human antibody coated onto the Acceptor bead, leading to a false-positive signal.
The table below shows data from experiments performed to determine which bead pairs might be able to interact on their own, to produce a false-positive signal. Donor/Acceptor bead pairings that have a higher background signal in the absence of other assay components may cause problems in assay design. These are indicated in yellow and red in the table. Bead pairings indicated in yellow yielded moderate counts in the absence of other assay components. Pairings indicated in red gave particularly high signals and are not recommended.
Table 2. Bead pairings test. Donor/Acceptor bead pairings indicated in red aren't recommended due to strong interactions between conjugated reagents on the beads. Pairs indicated in yellow may show higher background due to moderate interactions between the conjugated proteins/antibodies on the beads and could potentially be problematic in a given assay.
Note: This table refers to potential problematic interactions between the beads only. Other components being added to the assay should also be considered when designing your assay. For example, the table indicates a Protein A/anti-rabbit IgG bead pair would not interact on their own. However, if you are adding a rabbit IgG antibody to the assay, a single rabbit IgG antibody could bridge a Protein A Donor bead and anti-rabbit IgG Acceptor bead, leading to a false positive signal. This configuration could be used in very specific cases (for example, to titrate an anti-rabbit antibody), but should be generally avoided.
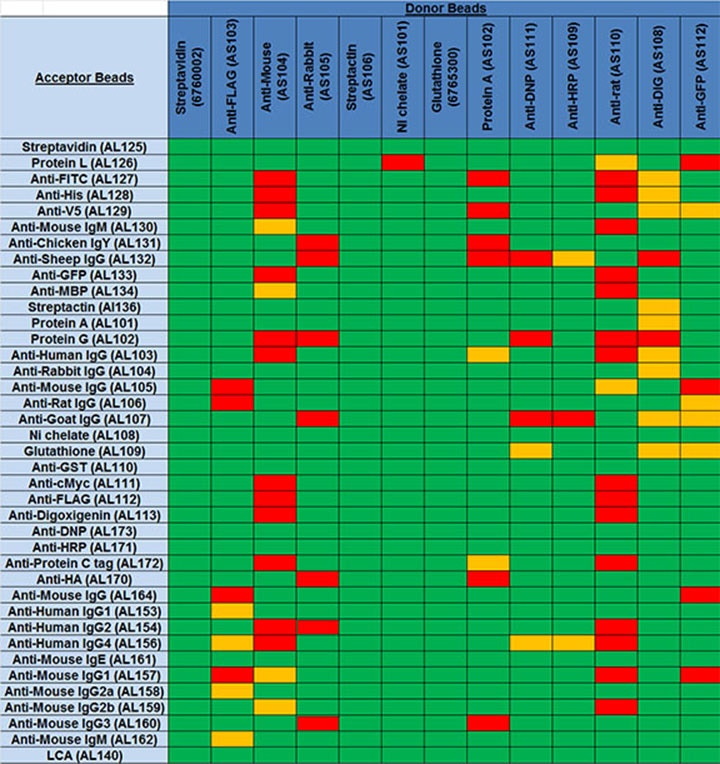
Bead pair chart
Buffer interference (by bead type)
Each bead product may be more or less susceptible to various components commonly found in biological buffers. For example, nickel chelate beads are susceptible to interference from EDTA because EDTA can chelate the nickel on the bead, rendering the bead unable to bind to His-tagged proteins. However, a streptavidin-coated bead is not as susceptible to interference from EDTA. Click the links below to view bead-specific interference data for common buffer components. Keep in mind that even if a particular bead is not susceptible to interference from an indicated buffer component, other reagents in your assay (proteins, antibodies, etc.) may be sensitive to the compound indicated - these interferences will be assay-specific, rather than bead specific. The information in the tables linked below is provided for guidance only.
- Streptavidin-coated AlphaLISA Acceptor beads
- Strep-Tactin®-coated beads
- Anti-6X His antibody-coated AlphaLISA Acceptor beads
- Nickel chelate AlphaLISA Acceptor beads
- Glutathione AlphaLISA Acceptor beads
- Anti-FITC antibody-coated AlphaLISA Acceptor beads
- Anti-FLAG antibody-coated Alpha Donor beads
- Anti-V5 antibody-coated AlphaLISA Acceptor beads
- Anti-GFP antibody-coated AlphaLISA Acceptor beads
- Anti-maltose binding protein (MBP) antibody-coated AlphaLISA Acceptor beads
- Protein A-coated Donor beads
- Protein L-coated AlphaLISA Acceptor beads
- Anti-mouse IgG antibody-coated Donor beads
- Anti-mouse IgM antibody-coated AlphaLISA Acceptor beads
- Anti-sheep IgG antibody-coated AlphaLISA Acceptor beads
- Anti-rabbit IgG antibody-coated Donor beads
- Anti-chicken IgY antibody-coated AlphaLISA Acceptor beads
Buffer interference (by buffer component)
This section provides the same information as in the previous section but sorted by buffer component. Each table indicates interference from a given compound, across each bead product. Click the links below to view compound-specific interference data for each Alpha Toolbox bead product. Keep in mind that even if a particular bead is not indicated as being susceptible to interference from a specific buffer component, other reagents in your assay (proteins, antibodies, etc.) may be sensitive to the compound listed - these interferences will be assay-specific, rather than bead specific. The information in the tables linked below is provided for guidance only.
- Glycerol
- DMSO
- Gelatin
- CHAPS
- Deoxycholate
- SDS
- Triton X-100
- Tween 20
- TRIS pH 8.0
- 2-mercaptoethanol
- DTT
- EDTA
- EGTA
- IBMX
- Imidazole
- Mg2+ (MgCl2)
- Nitroprusside
- Urea
- Vanadate
- Citrate
- Adenosine (ATP)
- Glycine
- Sodium fluoride (NaF)
- S-adenosylmethionine (SAM)
- Roche cOmplete™ Protease Inhibitor
Buffer interference (by bead type
Q. Can I use the same antibody on both beads?
A. Yes. This works best when using a polyclonal antibody, though. It can work for a monoclonal antibody as well, but here you are going to detect only homodimers. The latter could be a nice strategy for monitoring aggregation or dimerization. However, if you are monitoring aggregation or dimerization, the Alpha assay won’t be “quantitative”, meaning that a single pair or a big aggregate of hundreds of protein will give the “same” signal (signal from one bead pair). As for how to associate the same antibody to the two beads, there are various possibilities. The most classic one is to use a biotinylated version of the antibody with streptavidin-coated Donor beads and direct conjugation of the unlabeled version of the antibody on Acceptor beads. Double indirect association of the antibody (for example, using Protein A Donor beads and anti-rabbit IgG Acceptor beads to capture the same rabbit IgG antibody on both sides) would be trickier as it is possible a single rabbit IgG antibody might bridge a Donor and Acceptor bead in the absence of any other reagents, and is currently being evaluated.
Q. Can I use the same bead coating for both the Donor beads and the Acceptor beads in my assay design?
A. Yes, it is possible to use the same tag on both sides. There are two risks associated with this approach. First, the tag must be unique on each protein (i.e., one tag per protein). For example, a poly-biotinylated protein would not be ideal to use in this format, as a single poly-biotinylated protein could potentially bridge a streptavidin-coated Donor bead and a streptavidin-coated Acceptor bead, leading to a false positive signal. Second, there is also the risk of detecting not only protein 1-protein 2 complexes, but also protein 1-protein 1 or protein 2-protein 2 complexes (homodimers). Controls are therefore very important - each protein should be tested in the assay individually, to ensure no significant signal comes from either protein forming homodimers.
For research use only. Not for use in diagnostic procedures.
The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.