Overview
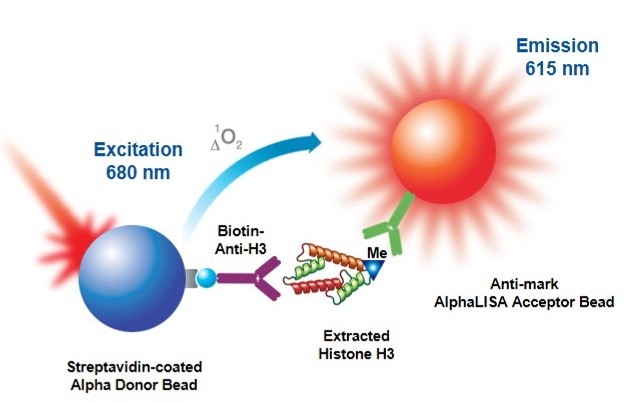
AlphaLISA™ epigenetic cellular detection kits were designed to detect endogenous epigenetic modifications on Histone 3 in a no-wash, all-in-one well format. The kits contain a proprietary buffer set which has been optimized to isolate cellular histones and allow for their detection using Alpha technology. In AlphaLISA assays, the level of epigenetic modification is detected by a mark specific antibody which is coated on an Alpha Acceptor bead. A biotinylated anti-Histone H3 antibody (specific to the C-terminus of Histone H3) binds to Histone 3 and is then captured by streptavidin-coated Alpha Donor beads. Upon laser irradiation of the bead complexes at 680 nm, short-lived singlet oxygen molecules produced by the Donor beads can reach the Acceptor beads in proximity to generate an amplified chemiluminescent signal at 615 nm. The intensity of light emission is proportional to the level of modification present.

Figure 1: AlphaLISA cell-based epigenetic assay principle. Histone H3 is extracted from cell lysates and detected with a biotinylated Anti-H3 antibody which binds streptavidin Donor beads and anti-mark antibody-coated AlphaLISA Acceptor beads specific to the particular modification (acetylated or methylated residue).
What do I need to run this assay?
Recommended reagents available from Revvity:
- AlphaLISA epigenetic cellular detection kits (see below section for product numbers)
- White CulturPlate™-384
- TopSeal-A™ adhesive sealing film
- An Alpha-capable plate reader, such as the EnVision™ multilabel plate reader
Optional (might be required for particular assays, see sections below):
- AlphaScreen™ SureFire® GAPDH Assay kit
- ATPlite™ Luminesence Assay
- AlphaLISA TruHits kit
- White OptiPlate™-384
- White CulturPlate-1536
- White ProxiPlate™-384 TC
Reagents available from other suppliers:
- Appropriate biotin-free culture medium for your cell line of interest
- Breathable sealing tape, sterile, available from Corning® (Product # 3345) or Sigma-Aldrich® (Product # CLS3345)
- Sodium butyrate (NaB), available from Sigma-Aldrich® (Product # B-5887-5G)
- Test compounds, as needed
Optional (might be required for particular assays, see sections below):
- Halt™ Protease Cocktail (100X), available from Thermo Scientific® (Product # 87786)
- Histone H3 Peptides (available from AnaSpec)
- Non-biotinylated peptides can be used to assess assay window and determine detection specificity (See Assay Development section below)
- Biotinylated peptides can be used as positive controls
Products and catalog numbers
We currently offer the following AlphaLISA Epigenetic Cellular Detection kits:
| Detected Modification | Description of Modification | Catalog Number |
|---|---|---|
| H3K4me2 | Histone 3 di-methylated on lysine 4 | AL716 |
| H3K9ac | Histone 3 acetylated on lysine 9 | AL714 |
| H3K27ac | Histone 3 acetylated on lysine 27 | AL720 |
| H3K27me3 | Histone 3 tri-methylated on lysine 27 | AL722 |
| H3K27me2-1 | Mono- and dimethylated-Histone H3 Lysine 27, in a cell-based assay | AL721 |
| H3K36me2 | Dimethylated-Histone H3 Lysine 36, in a cell-based assay | AL723 |
| H3K4 (unmodified) | Control: detection of unmodified Histone H3 Lysine 4, in a cell-based assay (H3K4) | AL719 |
| H3K79me2 | Dimethylated-Histone H3 Lysine 79, in a cell-based assay | AL748 |
| H3K9me2 | Dimethylated-Histone H3 Lysine 9, in a cell-based assay | AL717 |
The above kits include AlphaLISA anti-mark Acceptor beads, Streptavidin-coated Donor beads, Biotinylated anti-Histone 3 (C-term) Antibody, and the Epigenetic Cell-Histone™ Buffer Set, which includes buffers for cell lysis, histone extraction, and detection. Custom beads may also be available upon request, and custom assay development services are offered through our OnPoint™ custom teams (contact information is at the bottom of this page).
Protocol-in-brief
Below is an overview of the Universal No-Wash protocol, which is suitable for both adherent and suspension cell lines. The cells are plated and incubated in the presence or absence of compound. Cells are lysed, histones are extracted and the mark is detected using AlphaLISA™ reagents. If you are using adherent cells and need to include a wash step, you may do so after treating cells with compound and before adding Cell-Histone™ Lysis Buffer. For more information on when a wash step may be needed for your assay, please see the Tips and FAQs section below.
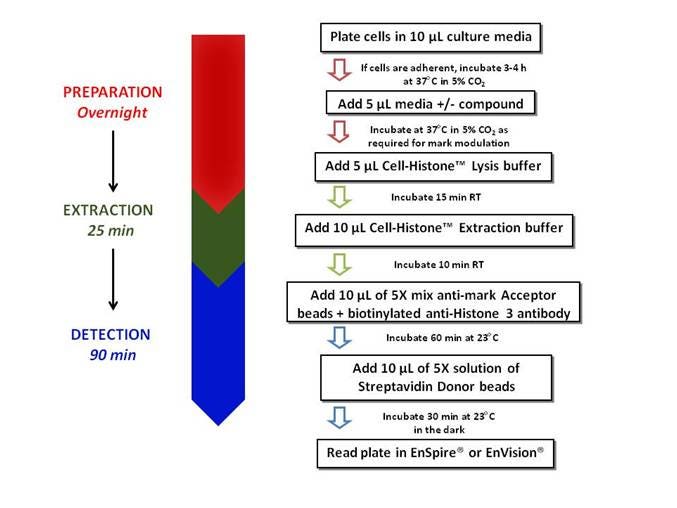
Figure 2. Workflow for AlphaLISA epigenetic cellular assay
Assay development
Assays should be optimized for your particular cell line, for each mark of interest. Examples for each mark can be found in the Technical Notes. Below is an example of recommended steps for assay optimization using the H3K9ac kit. In this example, two non-selective histone deacetylase (HDAC) inhibitors, sodium butyrate (NaB) and trichostatin A (TSA), were used to induce changes in the endogenous levels of H3K9ac in HeLa cells.
1. Detection of histone mark
Cell titration assay and western blot corroboration. We recommend setting up an experiment with increasing amounts of cell seeding densities followed by overnight treatment with NaB, or some other compound that should increase cellular levels of your mark of interest. You should observe a hook point as you increase cell number. The hook point may vary with cell lines. It is important to note that different cell lines exhibit different endogenous mark levels and different fold responses to treatment compounds. Therefore, in addition to a cell titration experiment, you may want to compare endogenous mark levels across several cell lines. Another important parameter for choosing the proper cell line for your assay is to confirm the mark modulation using a western blot.

Figure 3. A) HeLa cells were seeded at densities ranging from 100 to 10,000 cells/well in a 384-well CulturPlate™ and treated overnight with 20 mM sodium butyrate (NaB). B) For western blot analysis of H3K9ac mark modulation, 3 µg of cell lysate was separated by SDS-PAGE on a 10%-20% gradient gel. Following transfer to nitrocellulose, H3K9ac and total H3 were detected using antibodies, and western blots were revealed using alkaline-phosphatase-labeled anti-species secondary antibodies and Western Lightning™ CDP-Star® with Nitro-Block II™ Enhancer. C and D) AlphaLISA data and western blot analysis for H3K9ac modulation by 20 mM NaB in 3 different cell lines.
2. Detection specificity
experiment determines the kit's mark specificity in your cell line of interest.
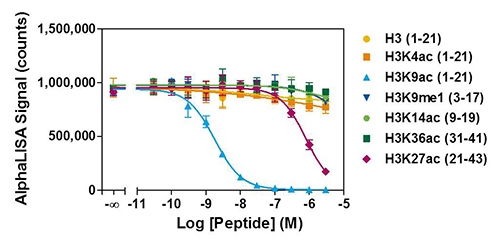
Figure 4. HeLa cells were seeded at a density of 2,000 cells/well and treated overnight with 20 mM NaB. Serial dilutions of Histone H3-derived peptides bearing various epigenetic marks were added to the wells at concentrations ranging from 30 pM to 3 μM just before the addition of AlphaLISA detection reagents. Only the H3K9ac peptide competed with high affinity for the interaction between the Acceptor beads and histone proteins with an IC50 value of 1.8 nM. The H3K27ac peptide yields an IC50 value of 778 nM.
3. Compound-induced modulation
This assay can be used to characterize inhibitors, or as part of assay validation using a reference inhibitor if you are screening. Increasing concentrations of inhibitor are tested at the cell seeding density determined from previous experiments. An EC50 for each compound can be derived from this data.
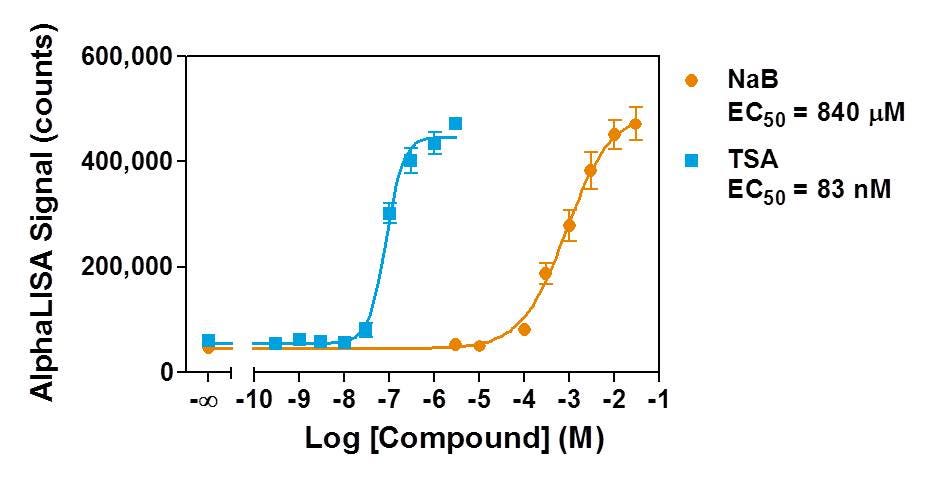
Figure 5. HeLa cells were seeded at a density of 2,000 cells/well and treated overnight with two non-selective HDAC inhibitors, TSA (from 300 pM to 3 μM) and NaB (from 3 μM to 30 mM), in medium containing 0.5% DMSO. TSA showed a 10,000-fold higher potency than NaB for increasing the general levels of H3K9ac marks in HeLa cells.
4. Assay variability
If you are screening, you may wish to perform a Z'-factor determination experiment as part of your assay validation. Two conditions are chosen that will generate a high and low signal (for example, with and without a particular concentration of HDAC inhibitor). In general, a Z'-factor above 0.5 is highly desirable for screening campaigns.
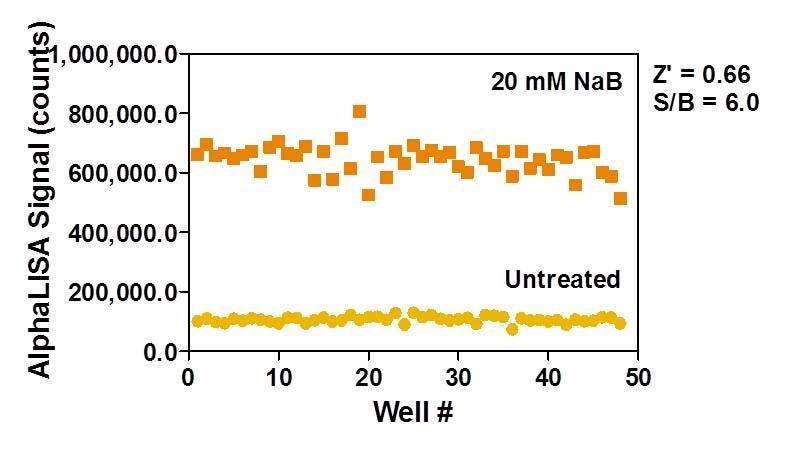
Figure 6. HeLa cells were seeded at a density of 2,000 cells/well and treated overnight with or without 20 mM NaB in medium containing 0.5% DMSO. The Z'-factor value compares sodium butyrate-treated and untreated cells.
Tips and FAQs
View our general tips and FAQs for working with Alpha. The following information is especially helpful when working with the epigenetic cellular detection kits:
- Biotin present in cell culture medium may interfere with the binding of biotinylated antibody to Streptavidin Donor beads. Use biotin-free culture medium when following the Universal Protocol. Most cells will not be affected by overnight incubation in biotin-free medium. If longer incubation is required, reduce biotin in culture medium by combining biotin-containing and biotin-free medium to find a condition not affecting cell growth or the Alpha signal. Alternatively, remove the medium and follow the Wash Protocol.
- Evaporation can be problematic with cells cultured in microtiter plates. For overnight incubations it is recommended to add warm PBS or sterile water to unused wells. For longer incubations, a sterile breathable sealing tape can be further added to the plate along with the lid.
- Additional kit-specific recommendations and troubleshooting tools can be found on the lot-specific technical data sheets.
Q. When do I need to follow the wash protocol?
A. The wash protocol can only be performed on adherent cells. A wash step is recommended if you are using biotin-containing media such as RPMI 1640 or if your test compound contains more than the recommended concentration of DMSO (found on the technical data sheet). If increased analytical sensitivity is desired, a wash step may also be helpful in increasing signal and assay window. Certain assays might require compounds that may interfere with mark detection using AlphaLISA. To test this, compounds should be incubated after the Histone Extraction step, but before the addition of the detection reagents. If a decrease in signal is observed, it can only be attributed to non-specific interference caused by the compound, and not to specific mark modulation. Another method to assess compound interference is to use our AlphaLISA TruHits kit. If it is determined that an interfering compound is present, the wash protocol should be followed.
Q. Does the lysis buffer contain protease inhibitors and DNases?
A. While we cannot disclose the components of our Cell-Histone™ buffers, the lysis buffer does not contain any protease inhibitors or DNases, as we found their presence did not improve assay performance for the conditions tested. However, you may add protease inhibitors to the lysis buffer if desired. In some cases when bulk cell lysates are being used, the abundance of genomic DNA can increase sample viscosity and variability. In these instances, DNase treatment may be beneficial. Because the lysis buffer contains ions essential for DNase activity, the enzyme can be added directly to the lysis buffer. Alternatively, DNase treatment may be added as an additional step after cell lysis.
Q. What if my mark of interest (H3K27me3) has no known modulators?
A. We have found that cell treatment with 20 mM sodium butyrate causes a detectable increase in the H3K9ac, H3K4me2, and H3K27ac marks. For these marks, sodium butyrate can be used as an effective modulation control. In cases such as the H3K27me3 mark where sodium butyrate treatment is unsuccessful, we recommend comparing two cell lines that display different endogenous levels of H3K27me3 such as the B cell lymphoma cell lines OCI-LY-19 and SU-DHL-16. For more information on assay development with the H3K27me3 kit, please download the Technical Note.
Q. What cell lines have you tested the AlphaLISA Epigenetic cell-based kits with?
A. We have validated the H3K9ac, H3K4me2, and H3K27ac kits with HeLa, Jurkat, HEK293 T, OCI-LY-19, and SU-DHL-6 cell lines. For the H3K27me3 kit, we have validated the B cell lymphoma cell lines OCI-LY-19 and SU-DHL-6. If you wish to use any of the kits on other cell lines, follow the Assay Development steps above.
Q. Can I stop the assay and freeze my cells at some point?
A. Yes. Frozen cell lystes can be prepared by following the normal procedure and freezing after the extraction step. However, there are some considerations:
For storage exceeding 1 week, add Protease inhibitors to the Cell-Histone Lysis buffer. Cocktails such as Halt’s prevent histone degradation that occurs upon longer-term storage.
- No need for snap-freezing on liquid nitrogen. Simply put the CulturPlates (with TopSeal) in the -20C freezer.
- For long term storage (more than one month) it is recommended to store the plates at -80C
- Thaw lysates at room temperature
- Lysates are also compatible with SDS-PAGE and Western-blotting
Q. What can I use as a positive control to validate kit reagents?
A. In addition to peptides from AnaSpec, there are several additional commercially available products which can be used with the epigenetic cell-based detection kits to both validate the kit reagents and serve as troubleshooting tools. Active Motif® sells recombinant Histone H3 proteins for the H3K4me2, H3K9ac, and H3K27me3 marks. Additonally, both untreated and sodium butyrate-treated HeLa cell extracts can be purchased from Active Motif® for the H3K4me2, H3K9ac, and H3K27ac marks.
Q. What can I use to normalize my results?
A. Results can be normalized by seeding cells in separate wells and using one of the following methods: AlphaScreen™ SureFire® GAPDH assay, or evaluation of cell density using imaging microscopy.
Q. Can I design my own cell-based assay for an epigenetic mark not currently offered as part of an AlphaLISA kit?
A. If you want to use AlphaLISA, please contact your Revvity specialist or Technical Support to discuss your mark of interest.
For research use only. Not for use in diagnostic procedures.
The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.