
Overview
Post-translational modifications of histones and other proteins are increasingly recognized as key contributors to the complex network of cellular regulation. Acetylation and methylation of histone proteins act to regulate gene activation at the chromosomal level. Acetylated histones tend to lead to gene activation, whereas methylated histones tend to be associated with gene inactivation. The enzymes that mediate these histone modifications have become attractive targets for drug discovery, and specific and sensitive assays are needed for screening and characterizing potential inhibitors of these enzymes. These enzyme classes include histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltrasferases (HMTs), and histone demethylases. The high sensitivity, dynamic range, scalability and resistance to fluorescent interference of AlphaLISA™ sandwich assays make them an attractive option for such assays. In addition, the ability of Alpha assays to measure interactions over larger (200 nm) distances makes them interesting for analysis of nucleosomes or other protein complexes.
The addition and removal of small ubiquitin or ubiquitin-like proteins from target proteins provides another level of regulating protein activity post-translationally. The diversity of proteins involved in the ubiquitin ligation pathway (the E1, E2, and E3 classes) and the large number of de-ubiquitinases (DUBs) suggest that this regulatory mechanism is a promising area in which to find specific drug targets. A general AlphaLISA sandwich assay format can be applied to many ubiquitinated proteins.
Additional post-translational modifications including phosphorylation, SUMOylation, and ADP-ribosylation can also play roles in transcriptional regulation, DNA repair, DNA replication, and chromosomal structure.
Assays
AlphaLISA epigenetic toolbox reagents
AlphaLISA™ epigenetics toolbox reagents were designed for the detection of specific methylation and acetylation marks on histone peptide substrates. In the Alpha assay, a biotinylated histone peptide substrate is used in a biochemical enzymatic reaction with an appropriate histone methyltransferase (HMT) or histone acetyltransferase (HAT) enzyme, using unlabeled S-adenosylmethionine (SAM) or Acetyl-CoA as a cofactor. When the enzymatic reaction is stopped, the level of methylation or acetylation is determined using antibody-coated AlphaLISA Acceptor beads and streptavidin-coated Alpha Donor beads. The streptavidin Donor beads capture the biotinylated substrate, and the antibody on the AlphaLISA Acceptor beads recognizes a specific mark on the substrate (acetylated Lysine 9 on Histone H3, di-methylated Lysine 9 on Histone H3, etc.). If the appropriate acetylation or methylation state exists on the substrate, the substrate will bring the Donor and Acceptor beads into proximity. Upon laser irradiation of the bead complexes at 680 nm, short-lived singlet oxygen molecules produced by the Donor beads can reach the Acceptor beads in proximity to generate an amplified chemiluminescent signal at 615 nm. The intensity of light emission is proportional to the level of biotinylated substrate modification.

Figure 1. Assay principle for AlphaLISA toolbox reagents epigenetics assay. Methylation assay using a biotinylated peptide substrate, S-adenosyl methionine, streptavidin Donor beads and anti-methyl antibody-coated AlphaLISA Acceptor beads.
AlphaLISA epigenetic cellular kits
AlphaLISA epigenetic cellular detection kits were designed to detect endogenous epigenetic modifications on Histone 3 in a no-wash, all-in-one well format. The kits contain a proprietary buffer set which has been optimized to isolate cellular histones and allow for their detection using Alpha technology. In AlphaLISA assays, the level of epigenetic modification is detected by a mark specific antibody which is coated on an Alpha Acceptor bead. A biotinylated anti-Histone H3 antibody (specific to the C-terminus of Histone H3) binds to Histone 3 and is then captured by streptavidin-coated Alpha Donor beads. Upon laser irradiation of the bead complexes at 680 nm, short-lived singlet oxygen molecules produced by the Donor beads can reach the Acceptor beads in proximity to generate an amplified chemiluminescent signal at 615 nm. The intensity of light emission is proportional to the level of modification present.
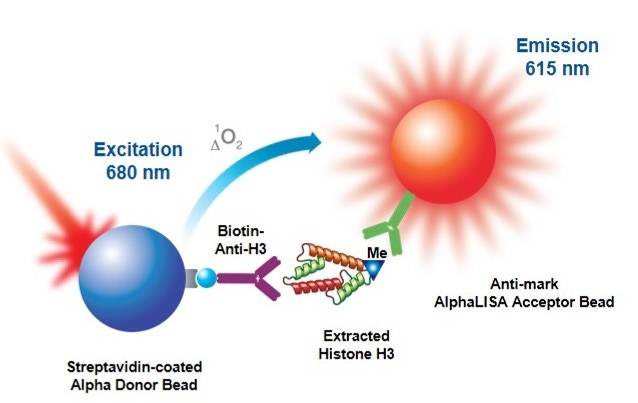
Figure 2. AlphaLISA cell-based epigenetic assay principle. Histone H3 is extracted from cell lysates and detected with a biotinylated Anti-H3 antibody which binds streptavidin Donor beads and anti-mark antibody-coated AlphaLISA Acceptor beads specific to the particular modification (acetylated or methylated residue).
Creating your own AlphaLISA epigenetic assay
View more information on creating other types of epigenetic assays using Alpha technology. In addition to the acetylation and methylation assays mentioned above, other types of post-translational modification detection assays have been created using Alpha reagents to detect specific epigenetic marks on both peptide and full-length histone and protein substrates.
Citations
View a brief list of citations describing Alpha assays for post-translational modifications.
For research use only. Not for use in diagnostic procedures.
The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.




























