Overview
Post-translational modifications of histones and other proteins are increasingly recognized as key contributors to the complex network of cellular regulation. In this regard, acetylation and methylation of histone proteins regulate gene expression at the chromatin level. Acetylated histones are generally associated with actively transcribed genes, whereas its methylation could lead to either gene activation or inactivation, depending on several contextual factors. The enzymes that mediate these histone modifications have become attractive targets for drug discovery, and specific and sensitive assays are needed for screening and characterizing potential modulator molecules. These enzyme classes include histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), and histone demethylases (HDMs). The high sensitivity, wide dynamic range, easy scalability, and resistance to fluorescent interference of AlphaLISA™ epigenetic toolbox reagents make them an attractive option for such endeavor. In addition, the ability of Alpha assays to measure interactions over large distances (200 nm) makes them interesting for the analysis of nucleosomes or other large protein complexes. A histone modification assay would most likely use a sandwich assay format in which one antibody recognizes the post-translational modification and a second antibody is used to capture the histone protein or complex. A simpler alternative to this would be to use a biotinylated peptide in place of the full-length histone protein as substrate for the enzymatic modification.

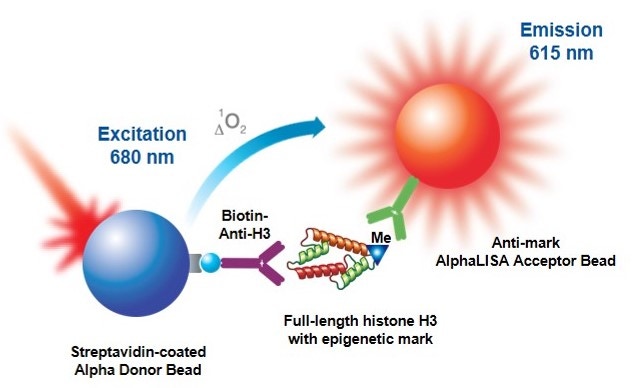
Figure 1. Detection of histone methylation in Alpha format, using a biotinylated antibody that recognizes the histone protein, a streptavidin-coated Donor bead, and an anti-mark antibody-coated Acceptor bead.
The modification of histones leads to chromatin regulation through interactions with a variety of chromatin-associated proteins. These interactions provide new therapeutic targets with the potential to have specific effects on chromatin expression. AlphaScreen and AlphaLISA assays can also be configured to detect the binding of modified histones to a variety of chromatin-associated proteins. A simple assay format could employ Donor beads linked to a biotin-tagged histone peptide bearing the modification of interest, and Acceptor beads linked to a GST-tagged chromatin binding protein (Quinn, 2009).
The addition and removal of ubiquitin or ubiquitin-like proteins from target proteins provides another level for the post-translation regulation of protein activity. The diversity of proteins involved in the ubiquitin ligation pathway (the E1, E2, and E3 classes) and the large number of de-ubiquitinases (DUBs) point to a promising area in which to find new drug targets. A general AlphaLISA sandwich assay format can be applied to many ubiquitinated proteins. This would include the capture of a tagged protein target by the Acceptor beads and a biotinylated antibody directed against ubiquitin or a directly biotinylated ubiquitin. Publications describing Alpha assays for ubiquitination and SUMOylation are available.
What do I need to run this assay?
Required reagents available from Revvity:
- AlphaScreen™ or AlphaLISA Acceptor beads that will associate with (or be conjugated to) an antibody recognizing the post-translational modification on your protein (or biotinylated peptide) substrate
- Biotinylated antibody to the target protein of interest (not required if a biotinylated peptide substrate is used)
- Alpha Streptavidin Donor beads used to bind to the biotinylated antibody (or biotinylated peptide substrate)
- Microplates - We recommend our 96-well 1/2 AreaPlates or our 384-well white OptiPlates™. Also see Microplate selection.
- TopSeal™-A adhesive plate seal for incubations
Instrumentation/equipment:
- A plate reader capable of reading Alpha assays (such as the EnVision™ or EnSight™ Multilabel Plate Reader)
Alpha products and catalog numbers
Complete listing of relevant Alpha products with catalog numbers.
Assay development
For assays involving post-translational modifications, you will most likely use a sandwich assay format where one antibody recognizes the post-translational modification and a second antibody that captures your target protein. In many cases, a biotinylated peptide can be used as a simpler, cheaper alternative substrate to the target protein. If your target is a tagged recombinant protein, you may set up the assay so that the target is captured by the expression tag rather than by an anti-protein antibody.
For more detailed information, please see Create your own Alpha assay.
Citations
View a brief list of citations describing Alpha assays for post-translational modifications.
Tips
- We recommend starting with 20 µg/mL Donor beads and 20 µg/mL Acceptor beads (final concentrations) in your detection reaction. If required, bead concentration can be titrated later in your assay development (in a cross-titration matrix, titrating each bead from 10 µg/mL to 40 µg/mL).
- The theoretical maximum capacity of a streptavidin-coated bead at 20 µg/mL of bead is 30 nM. Please note that this does not take into consideration the size of the biotinylated molecule that will associate with the bead. For example, you may find that a biotinylated antibody will saturate the bead at 2-3 nM, rather than at 30 nM. If you have saturated the bead, you may see a hook effect.
- The theoretical maximum capacity of an antibody-coated bead is 3-10 nM. Please note this is a theoretical number — you may be able to add more or less than 3-10 nM "antigen" to your assay without hooking, depending on how strong the antibody-antigen interaction is.
- The theoretical maximum capacity of a Ni-NTA or glutathione bead for a tagged protein is 3-30 nM, depending on the size of your protein. However, because the His-Ni-NTA interaction and the GST-glutathione interaction are weak (relative to a biotin:streptavidin interaction, for example), you may be able to use more than 30 nM tagged protein (possibly up to 100 nM or more) before saturating the bead.
For research use only. Not for use in diagnostic procedures.
The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.




























