
NEXTFLEX Small RNA Sequencing Kit V4

NEXTFLEX Small RNA Sequencing Kit V4




The NEXTFLEX® Small RNA-Seq Kit v4 enables labs to reduce bias in their Illumina® and Element Biosciences® small RNA sequencing workflows while leveraging a completely gel-free protocol, even with low input samples.
| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | Small RNA-seq |
The NEXTFLEX® Small RNA-Seq Kit v4 enables labs to reduce bias in their Illumina® and Element Biosciences® small RNA sequencing workflows while leveraging a completely gel-free protocol, even with low input samples.


NEXTFLEX Small RNA Sequencing Kit V4


NEXTFLEX Small RNA Sequencing Kit V4


Product information
Overview
The NEXTFLEX small RNA sequencing kit v4 uses patent-pending technology to provide a completely gel-free small RNA library preparation solution for Illumina and Element sequencing platforms.
- Completely gel-free protocol from purified miRNA or 1 ng of total RNA
- Automated on the Sciclone® G3 NGSx, Sciclone G3 NGSx iQ™, and the Zephyr® G3 NGS workstations
- Patent pending technology optimized for challenging samples such as plasma or serum, including exosomes
- Exceptional miRNA discovery
- Ready to sequence libraries in ~6 hours
- Multiplexing of up to 384 samples
- Quality assured through rigorous testing of every lot
Explore our Dharmacon™ miRNA modulation reagents to enhance or inhibit miRNA levels as part of your functional studies.
Additional product information
- Robustness across diverse sample types, from tissues to biofluids
- Streamlined workflow without gel cuts
- Fully automated - easy scalability from small to large studies
- Protocols available to select for small RNA species other than miRNA
We compared different vendors, and we found that the small RNA sequencing kit from Revvity was the one that performed best.
- Dr. Sören Franzenburg, Head of NGS platform, Kiel University
Gel-free small RNA library prep with reduced bias
The NEXTFLEX small RNA sequencing kit v4 uses patent-pending technology to provide a completely gel-free small RNA library preparation solution for Illumina sequencing platforms. Our approach to reducing bias involves optimization of the ligation reaction and the use of dimer reduction oligonucleotides. These changes result in decreased bias in comparison to standard protocols.
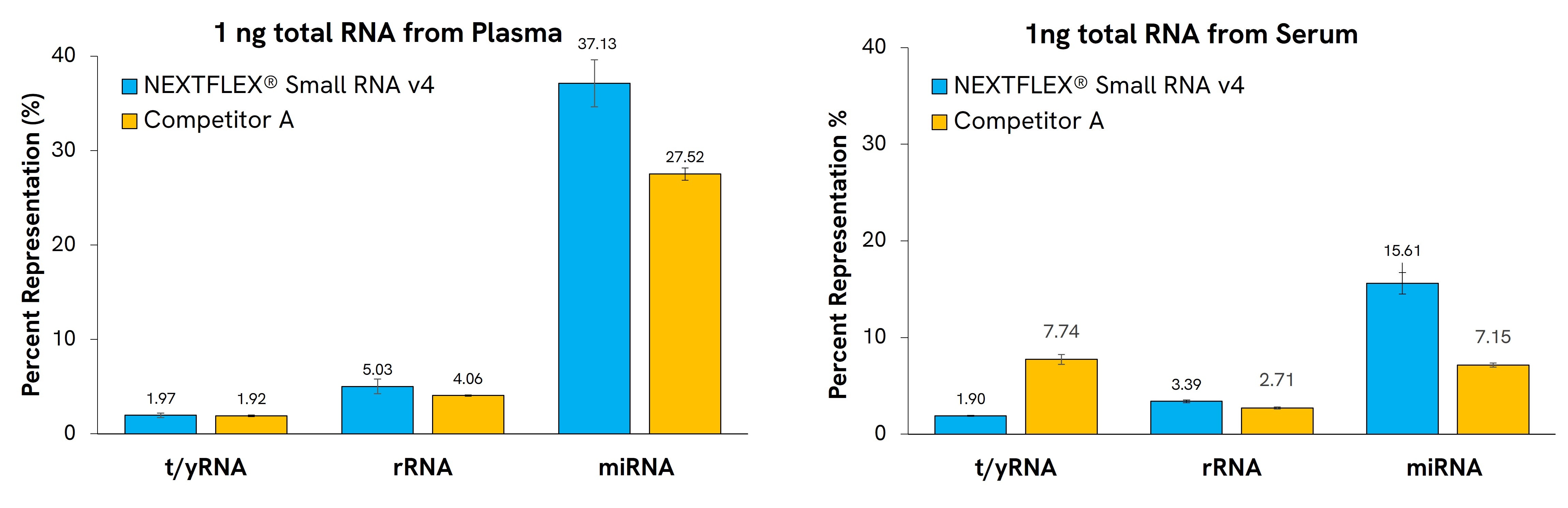
NEXTFLEX Small RNA-Seq Kit v4 allows robust preparation of libraries from diverse sample types in approximately 6 hours, with as little input as 1 ng of total RNA. Libraries prepared with this kit have a higher proportion of mapping reads and unique miRNAs than leading competitors. The kit contains tRNA and YRNA blockers to efficiently remove these species in biofluid samples (Figure 1 and 2).

Figure 1: Gel-free miRNA library prep from various biofluids
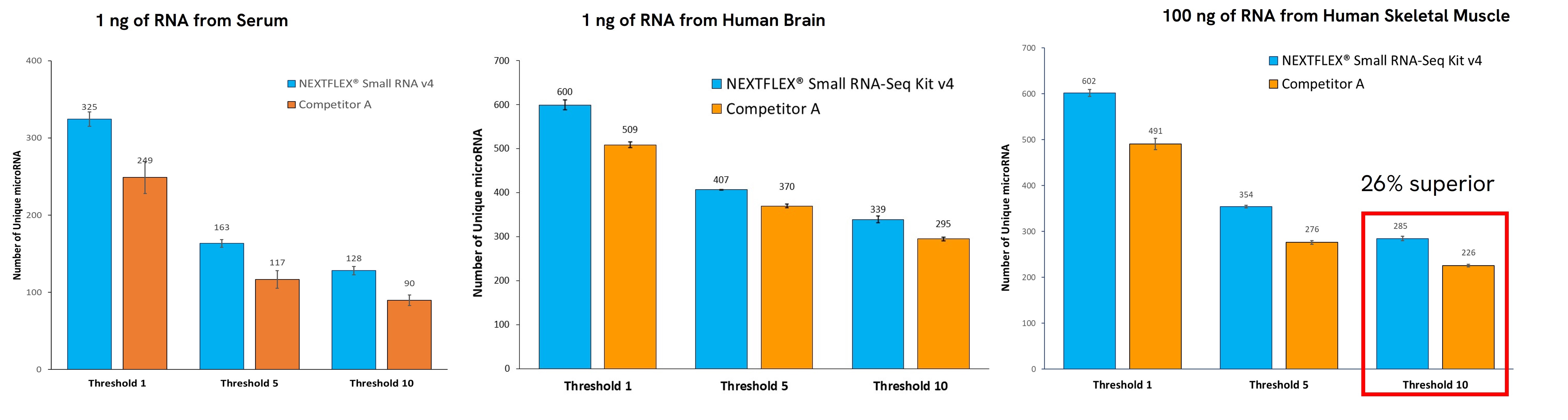
Figure 2: Gel-free miRNA library prep from exosomes
This convenient gel-free workflow incorporated in the NEXTFLEX Small RNA-Seq Kit v4 enables the construction of libraries from exoRNA. Using this workflow, researchers can achieve a high number of reads aligning to mature miRNA and low adapter dimers, even with the very low miRNA inputs characteristic of exosome samples (Figure 3).
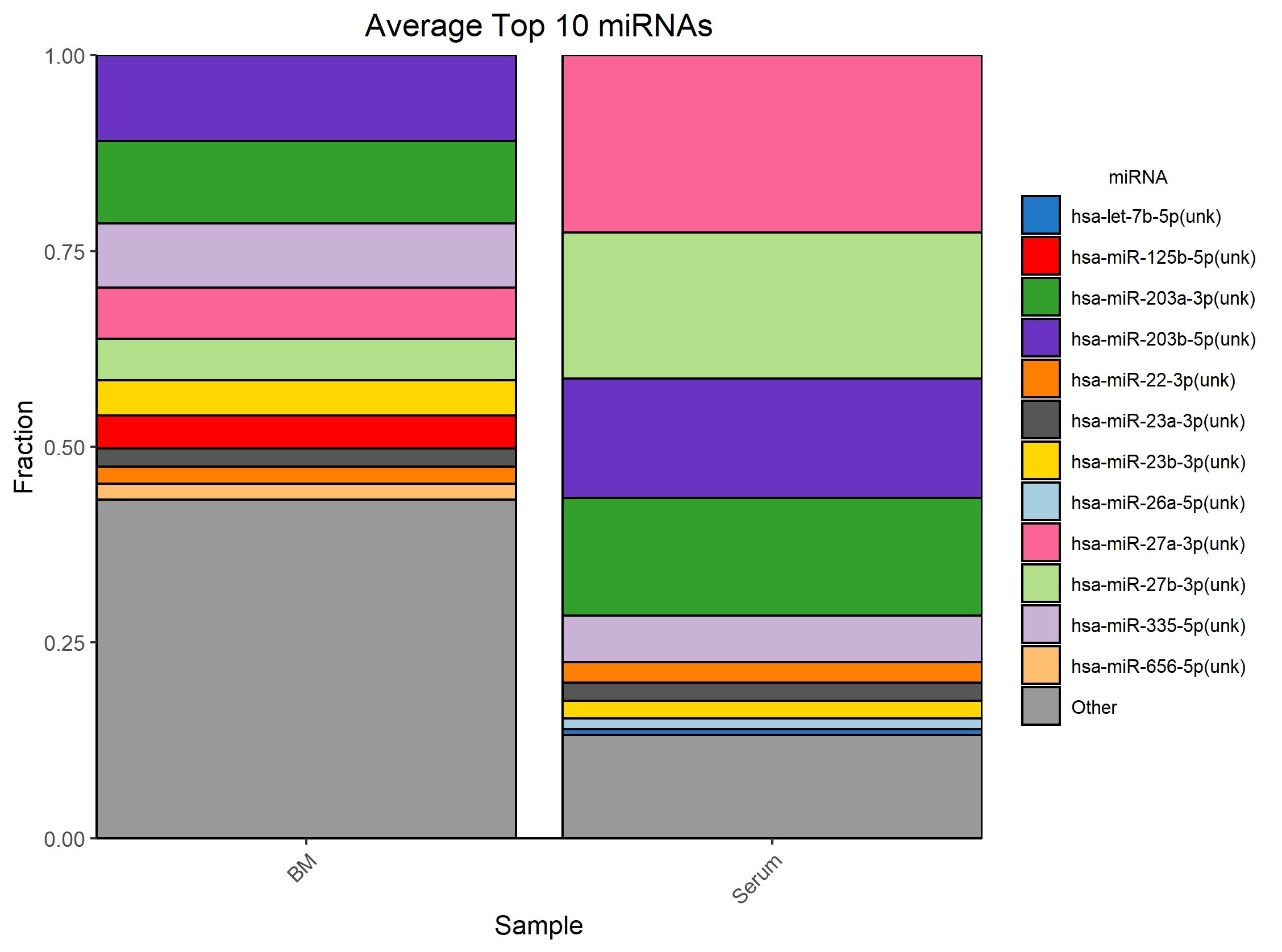
Figure 3: Average top 10 miRNA observed with exosomes from bone marrow and pooled serum samples.
Improved miRNA discovery
The performance of small RNA sequencing is heavily dependent on the workflow used. The NEXTFLEX® Small RNA Sequencing Kit v4 can identify a significantly higher number of unique miRNA species compared to other available solutions, even at low inputs (Figure 4). This comprehensive profiling is essential for understanding the full spectrum of miRNA expression in each sample. Detecting even the low-expressed miRNA present in the sample provides deeper insights into regulatory mechanisms and facilitates biomarker discovery.

Figure 4. Venn diagram showing that 45% more unique miRNA species were identified when using the NEXTFLEX® Small RNA-Seq Kit v4 than Competitor A kit. Input was 1 ng of RNA extracted from serum.
Ribosome Profiling (Ribo-Seq) Application
The NEXTFLEX Small RNA-Seq v4 Kit can also be used for ribosome profiling (Ribo-Seq), which analyzes ribosome-protected mRNA fragments to study translation. By adapting small RNA library preparation methods, researchers can gain insight into gene regulation and translation dynamics, making Ribo-Seq valuable in fields like gene expression, disease mechanisms, and drug discovery.
“We frequently use Revvity small RNA sequencing kits for Ribosome- and Disome-profiling. It offers a very streamlined library generation and allows for easy customization to adapt to experimental needs. With this, it consistently delivers good results. Additionally, the technical support from Revvity’s team is excellent and very helpful which further enhanced our very positive experience with this Revvity product.”
- Dr. Tobias Schmidt, CRUK Scotland Institute
Small RNA sequencing library prep optimized for automation
By eliminating all gel-based steps even for low input samples, the NEXTFLEX Small RNA-Seq Kit V4 is optimized for easy migration onto automated liquid handling platforms. This workflow is automated on the Sciclone G3 NGSx, Sciclone G3 NGSx iQ, and the Zephyr G3 NGS workstations
Illumina® and Element® small RNA sequencing multiplexing with UDI barcodes
Up to 384 UDI barcoded primers are included in each kit for multiplexing. These UDIs allow libraries to be run on any Illumina and Element Biosciences sequencing platform. These include the Illumina NextSeq® and Novaseq® sequencers and only require standard Illumina sequencing primers.
Maximize miRNA mapped reads with NEXTFLEX blood miRNA blockers
The NEXTFLEX blood miRNA blockers deplete common miRNA in blood and plasma such as miR-486-5p, miR-92a-3p and miR-451a. These constitute 50-70% of the small RNA present in those samples and usually are not of interest for researchers.
We have been using Revvity small RNA sequencing kits for a number of years, and we are satisfied with the results and the support received.
- Dr. Marcel van Herwijnen, Researcher/Lab Manager, Maastricht University
Listen to the Experts:
Listen to our webinar, "Exploring Circulating Extracellular Vesicles in Liquid Biopsies for Neurological Disorders," where Bessie Meechoovet, a Senior Research Associate from the Jensen lab at the Translational Genomics Research Institute (TGen), explains how cell-free biomarkers are used to enhance our understanding of neurodegenerative diseases.
The Jensen lab utilizes long-read single-cell and RNA transcriptomics data to identify minimally invasive biomarkers in biofluids. Ms. Meechoovet describes a method they've developed to enrich brain extracellular vesicles, aiding in the discovery of potential biomarkers.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Barcodes |
289 - 384
|
| Format |
Automation Friendly Volumes
|
| Product Group |
Small RNA-seq
|
| Shipping Conditions |
Dual Temperature
|
| Unit Size |
96 rxns
|
Citations
- Almeida, M.V., Blumer, M., Yuan, C.U. et al. Dynamic co-evolution of transposable elements and the piRNA pathway in African cichlid fishes. Genome Biol 26, 14 (2025). https://doi.org/10.1186/s13059-025-03475-z.
- Apostolov, A., Mladenović, D., Tilk, K. (2025) Multi-omics analysis of uterine fluid extracellular vesicles reveals a resemblance with endometrial tissue across the menstrual cycle: biological and translational insights, Human Reproduction Open, https://doi.org/10.1093/hropen/hoaf010.
- Chu, C. P., Nabity, M. B. (2025) Technical considerations and review of urinary microRNAs as biomarkers for chronic kidney disease in dogs and cats. Vet Clin Pathol. 2025; 00: 1-19. doi:10.1111/vcp.13412.
- Karere, G. M., Hsu, F.-C. Hepple, R. T. et al. (2025) MicroRNA signatures of VO2peak in older adult participants of the Study of Muscle, Mobility and Aging. bioRxiv 2025.01.08.631999; doi: https://doi.org/10.1101/2025.01.08.631999.
- Lill C. M., et al. (2025) EPIC4ND: European Prospective Investigation into Cancer and Nutrition follow-up for neurodegenerative diseases. medRxiv 2025.01.29.25321340; doi: https://doi.org/10.1101/2025.01.29.25321340.
- Marocco. F. (2025) The RNA binding protein PCBP2 is a regulator of miRNAs partition between cell and extracellular vesicles. [Doctoral dissertation, Sapienza University of Rome]. IRIS Uniroma1. https://iris.uniroma1.it/retrieve/6d5c8022-59e2-44ec-b8b6-eb7a6ea8cc6e/….
- Rossner, p., Javorkova, E., Sima, M. et al. Skin wound healing: the impact of treatment with antimicrobial nanoparticles and mesenchymal stem cells. (2025) Authorea. February 17, 2025. DOI: 10.22541/au.173977945.54128573/v1.
- Tahiri, I., Llana, S.R., Díaz-Castro, F. et al. AgRP neurons shape the sperm small RNA payload. Sci Rep 15, 7206 (2025). https://doi.org/10.1038/s41598-025-91391-4.
FAQs
-
What types of RNAs are included in the Small RNA library?
-
Do you have any recommendations for extraction of RNA for Small RNA sequencing?
-
Are the libraries produced by NEXTFLEX Small RNA Sequencing Kit stranded/directional?
-
Can I use total RNA, or do I have to isolate or enrich the sample for small RNA?
-
What happens if the quality of my RNA is very low?
-
How should my NEXTFLEX Small RNA libraries be trimmed?
-
What should I do if my Small RNA may not have 5' Phosphate and 3' OH groups?
-
How are the barcodes introduced in the NEXTFLEX Small RNA libraries?
-
What are the recommended read length and settings for sequencing?
-
What is the number of reads required for small RNA sequencing?
-
Do I have to use custom Illumina® sequencing primers for the libraries generated with NEXTFLEX Small RNA Sequencing Kit v4?
-
What do I need to do to run the NEXTFLEX Small RNA libraries on the AVITI™ sequencer from Element® Biosciences?
-
Are the Monarch™ Spin RNA Cleanup Kits (NEB) compatible with NEXTFLEX reagents for RNA library prep?
-
Do you have recommendations for analysis of small RNA sequencing data?
Resources
Are you looking for resources, click on the resource type to explore further.
This flyer describes the benefits of the NEXTFLEX® Small RNA-Seq Kit v4 kit
In this application note we describe a simplified, commercially available protocol encompassing exoRNA extraction and preparation...
NEXTFLEX® Small RNA-Seq Kit v4 has a streamlined, gel-free workflow delivering exceptional miRNA mapping and discovery rates even...
This poster illustrates the effieciency of the NEXTFLEX® Small RNA-Seq Kit v4 kit of discovering biomarkers.
In the rapidly evolving field of molecular biology, the extraction and analysis of small RNA from plasma have become pivotal for...


How can we help you?
We are here to answer your questions.






























