
HTRF Human and Mouse Total PARP1 Detection Kit, 10,000 assay points
HTRF Human and Mouse Total PARP1 Detection Kit, 10,000 assay points


This HTRF kit allows for the cell-based quantitative detection of Total PARP1.
| Feature | Specification |
|---|---|
| Application | Cell Signaling |
| Sample Volume | 16 µL |
This HTRF kit allows for the cell-based quantitative detection of Total PARP1.


HTRF Human and Mouse Total PARP1 Detection Kit, 10,000 assay points


HTRF Human and Mouse Total PARP1 Detection Kit, 10,000 assay points


Product information
Overview
PARP1 (Poly(ADP-ribose) polymerase 1) is a nuclear enzyme that plays a key role in DNA repair by utilizing NAD⁺ to synthesize poly(ADP-ribose) (PAR). Its overexpression in various cancers contributes to tumor progression. In cells with BRCA1 or BRCA2 deficiencies, PARP1 inhibition leads to apoptosis, making it a promising therapeutic target in cancer treatment.
Specifications
| Application |
Cell Signaling
|
|---|---|
| Brand |
HTRF
|
| Detection Modality |
HTRF
|
| Lysis Buffer Compatibility |
Lysis Buffer 4
|
| Molecular Modification |
Total
|
| Product Group |
Kit
|
| Sample Volume |
16 µL
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Target |
PARP1
|
| Target Class |
Phosphoproteins
|
| Target Species |
Human
Mouse
|
| Technology |
TR-FRET
|
| Therapeutic Area |
Oncology
|
| Unit Size |
10,000 assay points
|
How it works
Total PARP1 assay principle
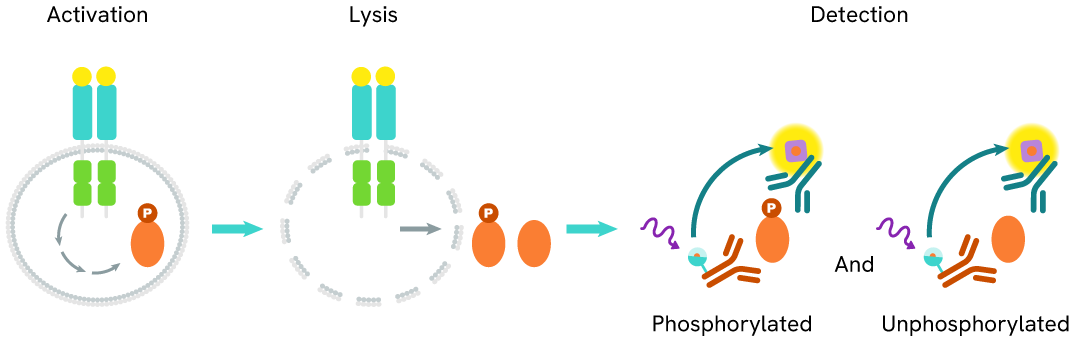
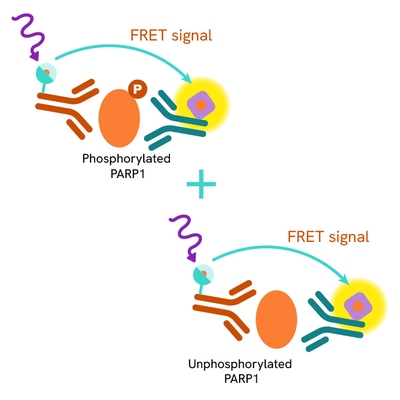
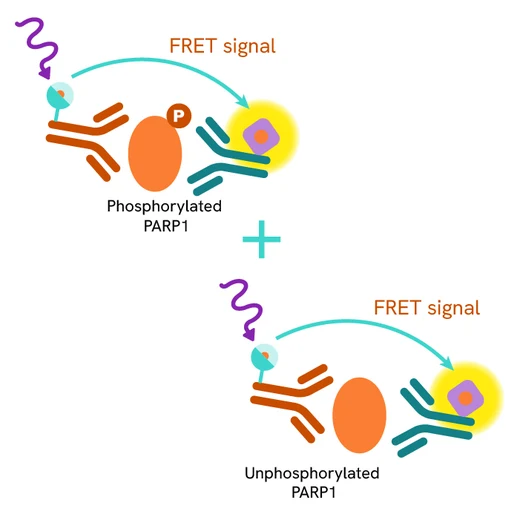
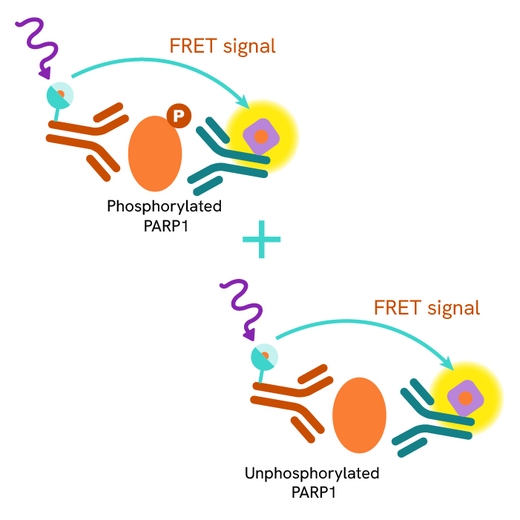
The Total PARP1 assay quantifies the expression level of PARP1 in a cell lysate. Unlike Western Blot, the assay is entirely plate-based and does not require gels, electrophoresis, or transfer. The assay uses two labeled antibodies, one coupled to a donor fluorophore and the other to an acceptor. Both antibodies are highly specific for a distinct epitope on the protein. In the presence of PARP1 this enables an immune-complex formation involving both labeled antibodies, and which brings the donor fluorophore into close proximity to the acceptor, thereby generating a FRET signal.
Its intensity is directly proportional to the concentration of total protein present in the sample, and provides a means of assessing the protein's phosphorylation state under a no-wash assay format.

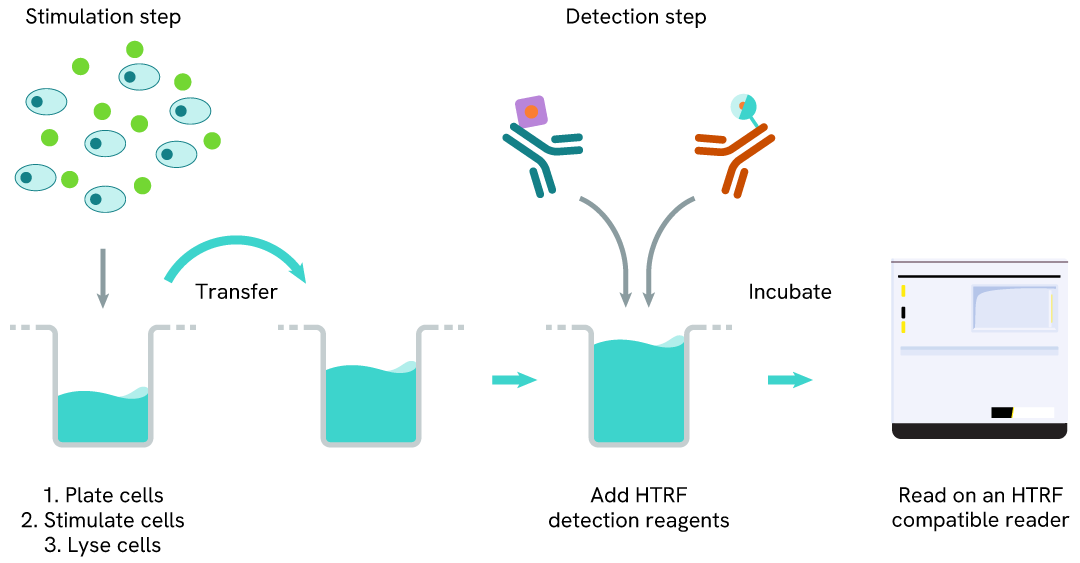
Total PARP1 two-plate assay protocol
The two-plate protocol involves culturing cells in a 96-well plate before lysis, then transferring lysates into a 384-well low volume detection plate before the addition of HTRF Total PARP1 detection reagents. This protocol allows for the cells' viability and confluence to be monitored.
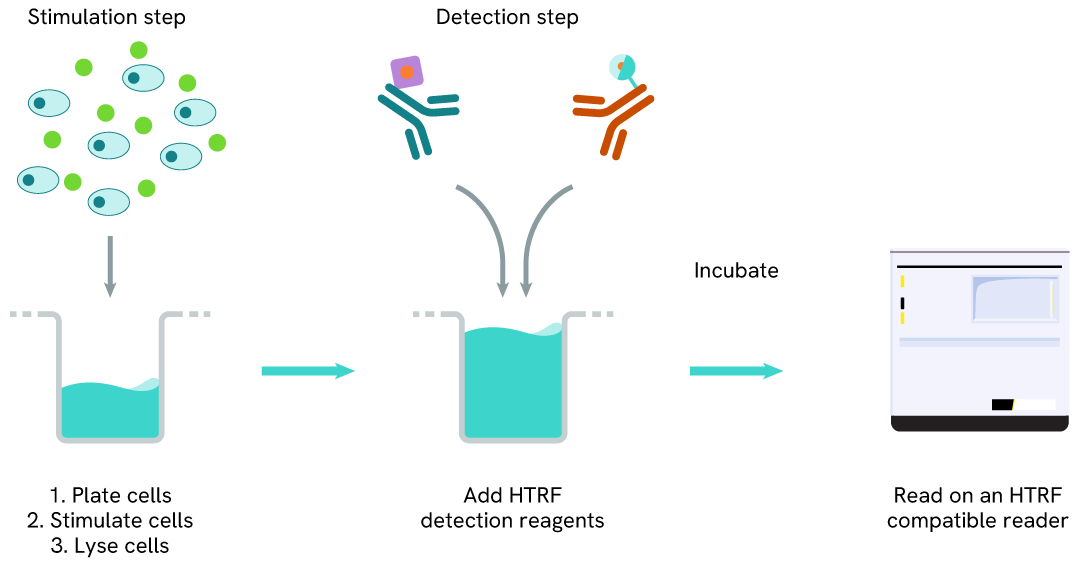
Total PARP1 one-plate assay protocol
Detection of Total PARP1 with HTRF reagents can be performed in a single plate used for culturing, stimulation, and lysis. No washing steps are required. This HTS designed protocol allows miniaturization while maintaining robust HTRF quality.
Assay validation
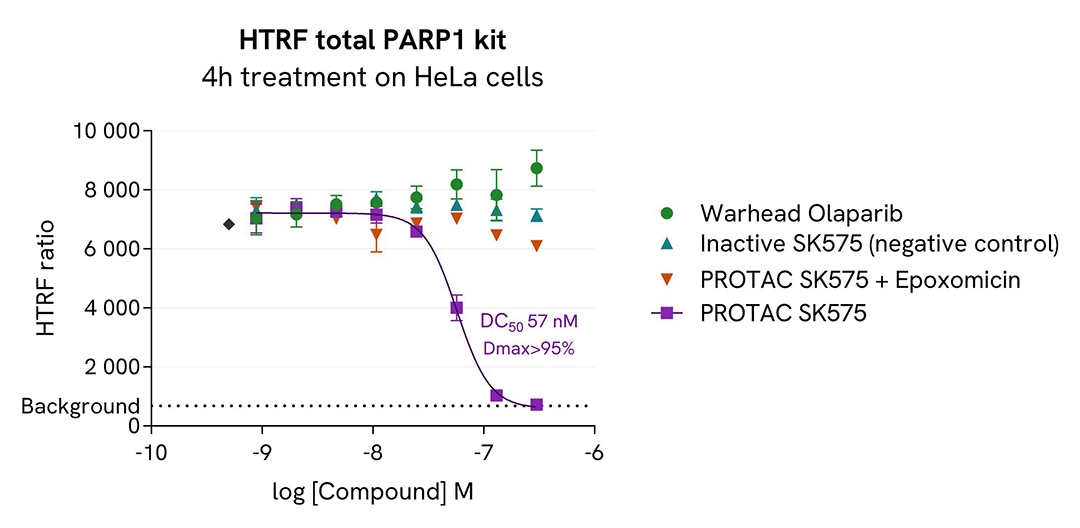
PROTAC® induced PARP1 degradation
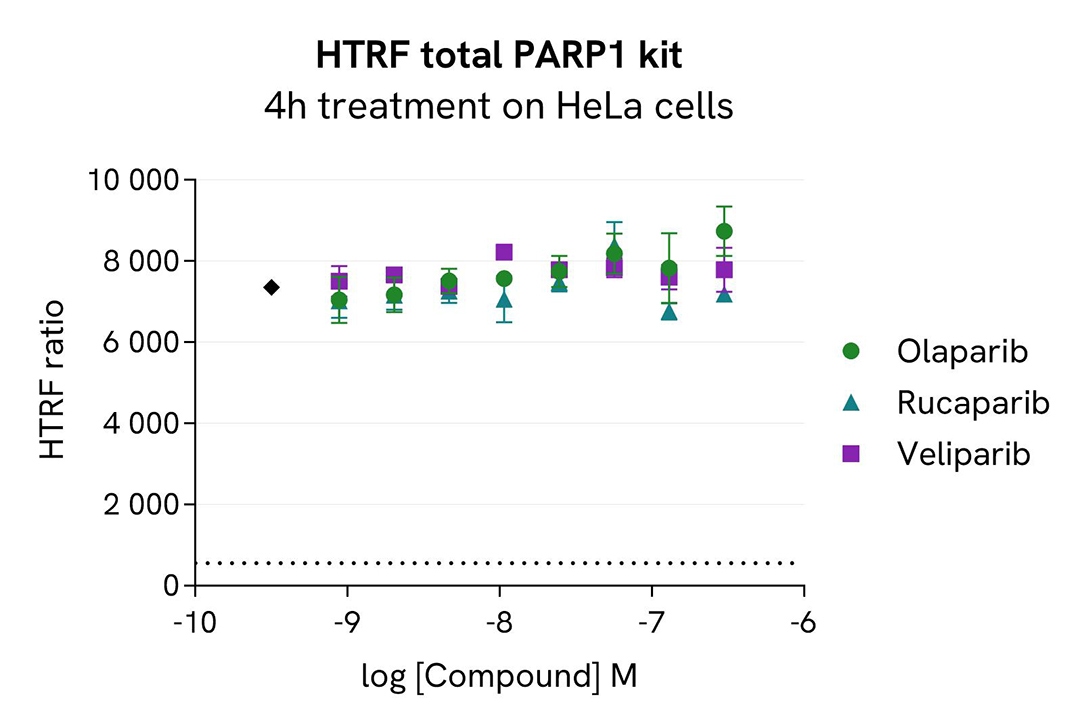
HeLa cells were cultured in a 96-well plate (25,000 cells/well) for 24h at 37°C, 5% CO2. The day after, the cells were first pre-treated for 1 hour with 1 µM Epoxomicin and then treated during an additional 3h-incubation with increasing concentrations of PROTAC® compound SK-575, which is a highly potent and specific PARP1 degrader compared to PROTAC® alone, the negative control (inactive), or inhibitors of PARP1 enzymatic activity (Olaparib, Rucaparib, and Veliparib).
Next, cell culture medium was removed and 50 µl of supplemented Lysis Buffer#4 (1X) were dispensed into each well. After cell lysis, 16 µL of lysates were transferred into a 384-well low volume white microplate and 4 µL of the HTRF PARP1 detection antibodies were added. The HTRF signal was recorded after an overnight incubation. Cell viability was also assessed using ATPlite 1step detection reagent (ATPlite 1step Luminescence Assay System, Cat # 6016736/1/9). No toxicity was observed (data not shown).
After 4 hours of treatment, the PROTAC® degrader triggered a dose-dependent decrease in PARP1 protein, whereas the negative control and inhibitors caused no changes, as expected. The HTRF signals, recovered in presence of the proteasome inhibitor epoxomicin, showed a degradation of PARP1 mediated by the ubiquitin proteasome system.
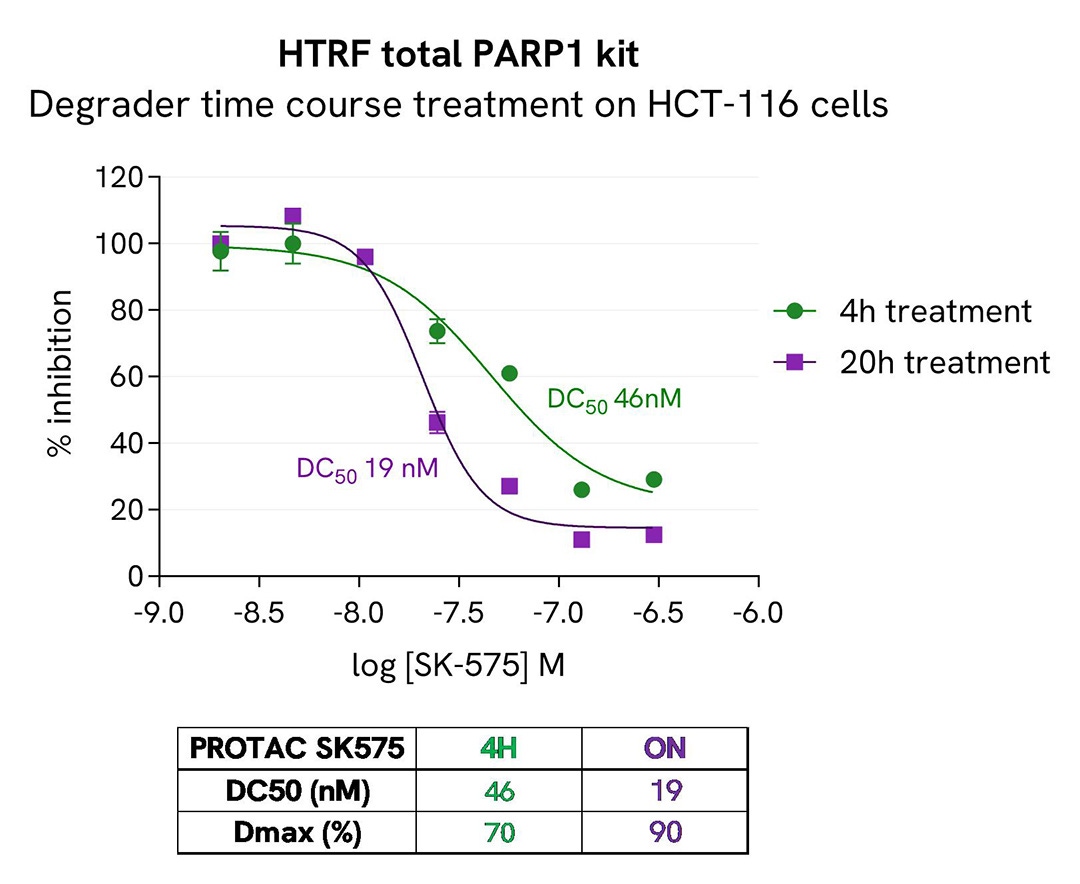
Dose-effect and time-course studies of a PARP1 PROTAC®
HCT-116 cells were cultured in a 96-well plate (25,000 cells/well) for 24h at 37°C, 5% CO2, and then treated 4h and 20h with increasing concentrations of PROTAC® compound SK-575, a highly potent and specific degrader of PARP1.
The cell culture medium was removed and 50 µl of supplemented Lysis Buffer#4 (1X) were dispensed into each well. After cell lysis, 16 µL of lysates were transferred into a 384-well low volume white microplate and 4 µL of the HTRF PARP1 detection antibodies were added. The HTRF signal was recorded after an overnight incubation.
The PROTAC degrader triggered a dose-dependent decrease in PARP1 protein with improved potency and efficacy after 20h of treatment. No cytotoxic effect was observed in presence of the compound, even after a 20h treatment (ATPlite 1step assay, data not shown).
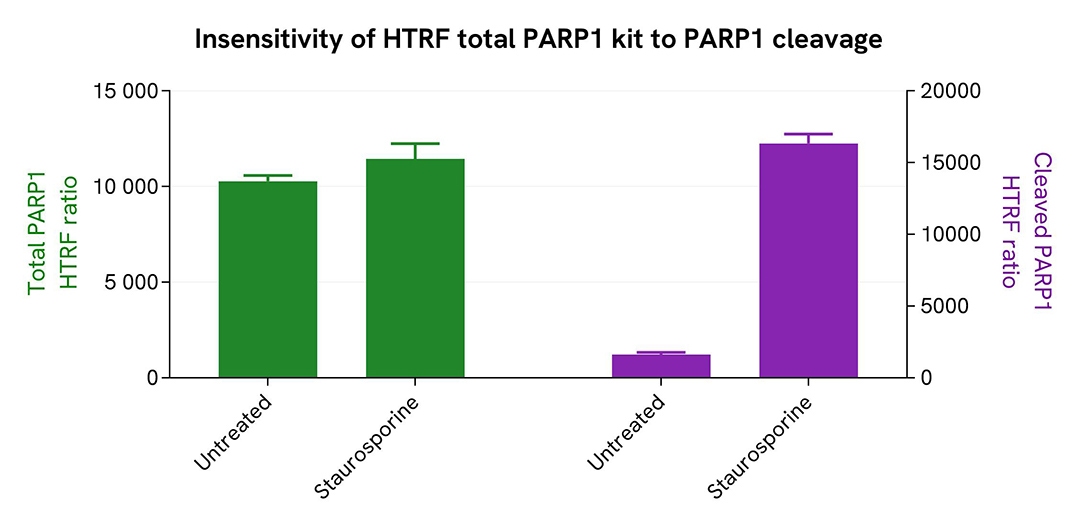
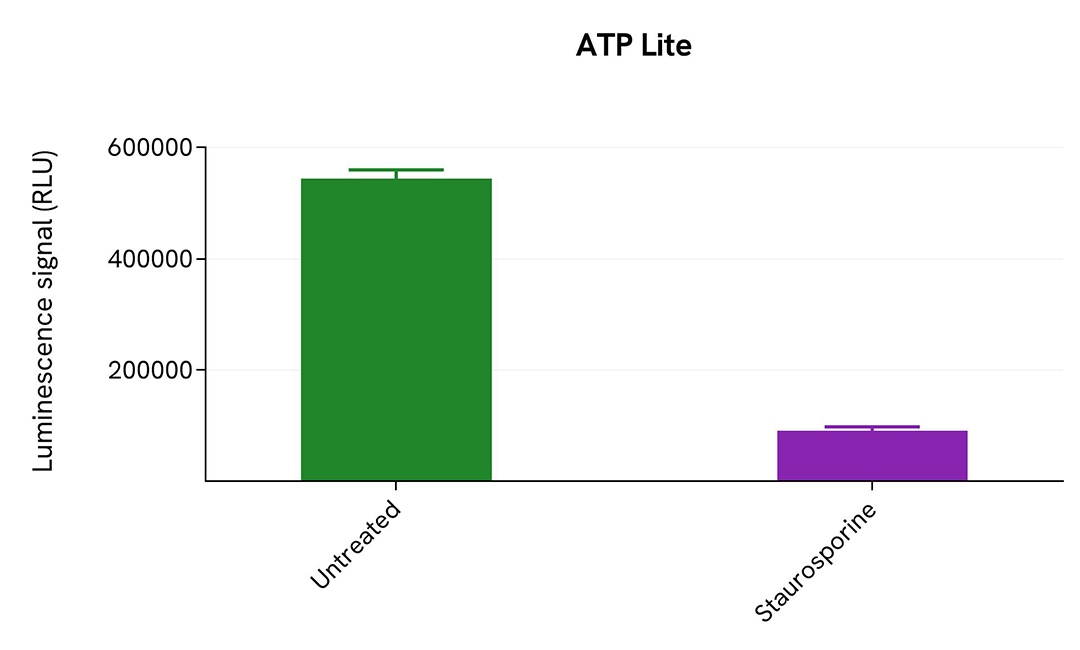
Total PARP1 is insensitive to cleavage
HeLa cells were plated in 96-well culture plates (25,000 cells/well) in complete culture medium and cultured overnight at 37°C, 5% CO2. The following day, the cells were treated with Staurosporine 3 µM for 4 hours, while the control was not treated. The medium was removed, and cells were lysed with 50 µL of supplemented lysis buffer #1 for 30 min at RT under gentle shaking.
16 µL of lysate were transferred into a 384-well sv white microplate, and 4 µL of the HTRF Cleaved PARP1 Asp214 (#64PARPEG) or Total PARP1 detection reagents were added. The HTRF signal was recorded for both assays after an overnight incubation.
Cell viability was also assessed by transferring 5 µL of the same lysate into an HTRF 96-well low volume white plate (Cat # 66PL96005/025/100), and then adding 25 µL of ATPlite 1step detection reagent (ATPlite 1step Luminescence Assay System, Cat # 6016736/1/9). The luminescence signal was measured after a 10-min incubation in the dark.
Staurosporine treatment triggered PARP1 cleavage through the induction of apoptosis, leading to cell death visible by a significant drop in ATPlite. Meanwhile, Total PARP1 remained unchanged.
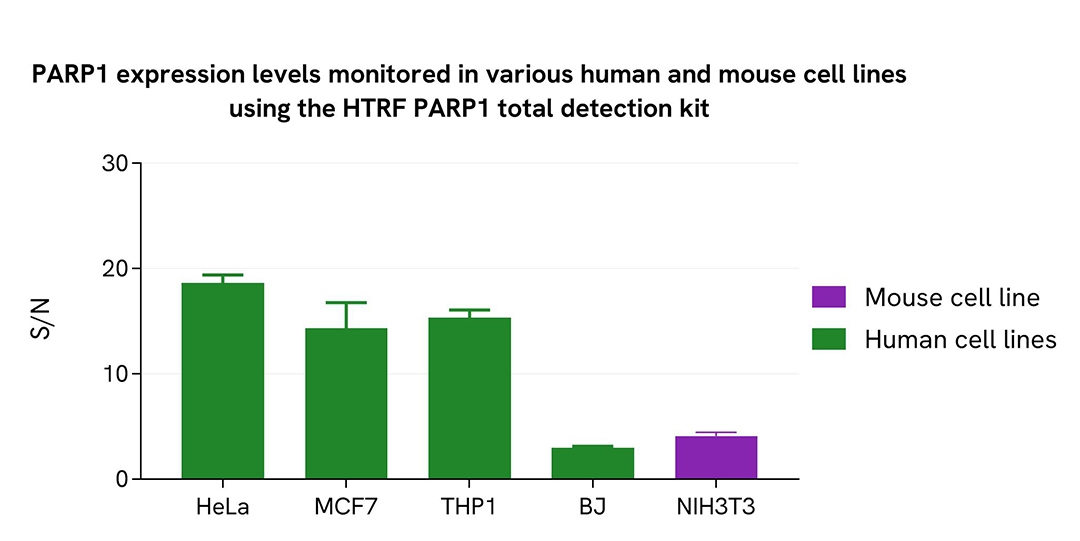
PARP1 expression levels monitored in various human and mouse cell lines
Adherent human cells HeLa (cervix), MCF7 (breast cancer), BJ (fibroblast), or suspension cells THP-1 (monocyte) and mouse cells NIH-3T3 (fibroblasts), were seeded at 50,000 cells/well in a 96-well microplate for 24 hours at 37°C, 5% CO2. After culture medium removal, the cells were lysed with supplemented lysis buffer #4 (1X).
The Total PARP1 level was assessed with the HTRF Total PARP1 kit. Briefly, 16 µL of cell lysate were transferred into a 384-well low volume white microplate before the addition of 4 µL of premixed HTRF detection reagents. The HTRF signal was recorded after an overnight incubation at RT.
The HTRF Total PARP1 assay efficiently detected Total PARP1 in various cellular models expressing different levels of the protein.
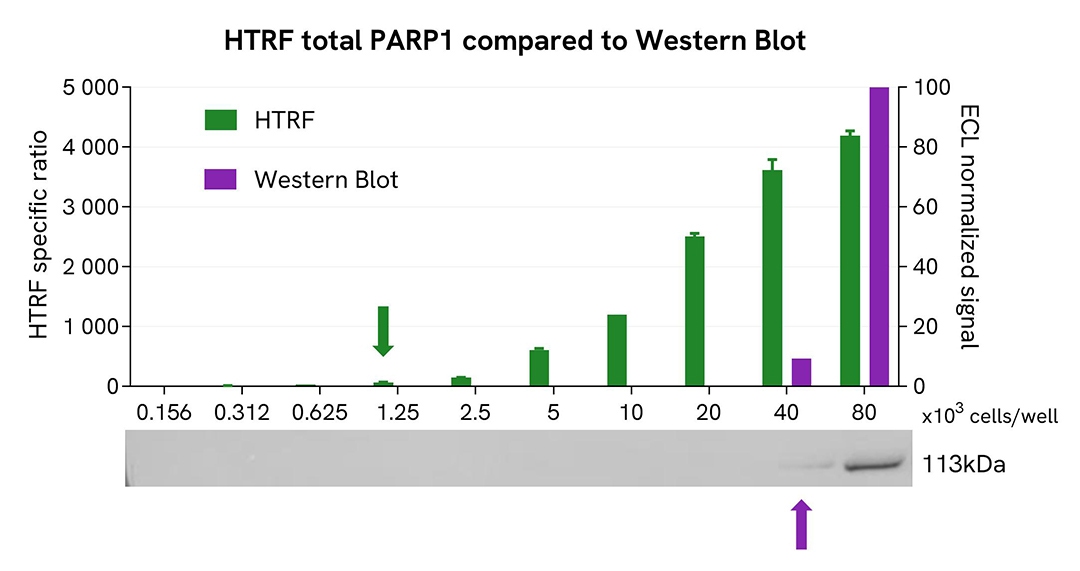
HTRF Total PARP1 assay compared to Western Blot
HeLa cells were grown in a T175 flask in complete culture medium at 37°C, 5% CO2, until 80% confluence. After a 48h incubation and cell medium removal, the cells were lysed with 3 mL of supplemented lysis buffer #4 (1X) for 30 minutes at RT under gentle shaking.
Serial dilutions of the cell lysate were performed using supplemented lysis buffer #4, and 16 µL of each dilution were transferred into a low volume white microplate before the addition of 4 µL of HTRF Total PARP1 detection reagents.
Equal amounts of lysates were used for a side-by-side comparison between HTRF and Western Blot.
In these conditions, the HTRF total PARP1 assay was 32-fold more sensitive than the Western Blot technique.
Simplified pathway
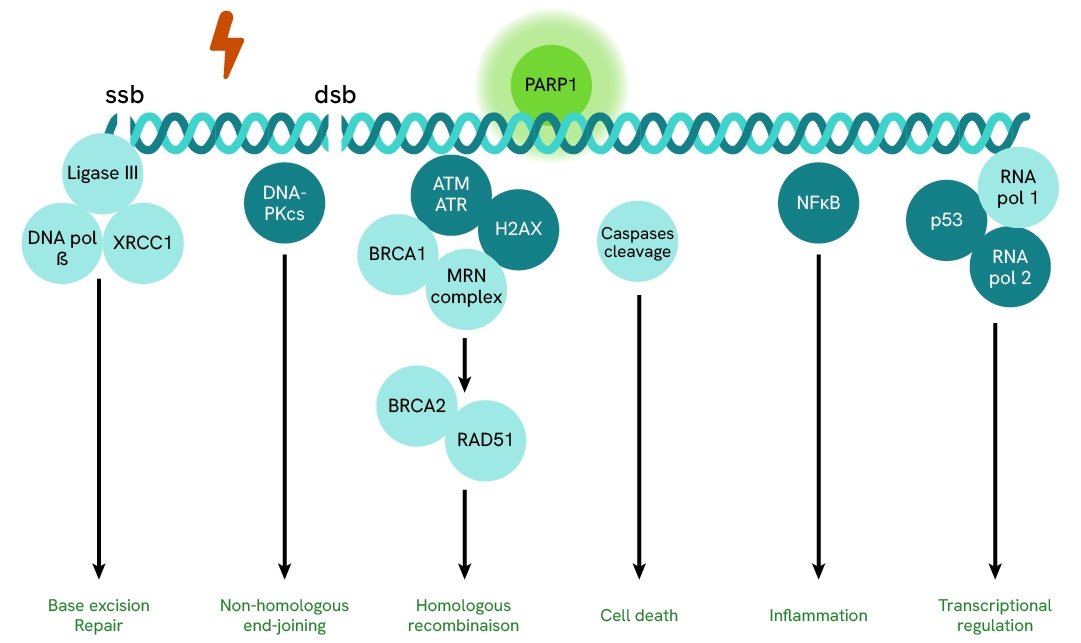
PARP1 signaling pathway
PARP1 (Poly(ADP-ribose) polymerase 1 ) is a key enzyme involved in multiple cellular processes. In the DNA damage response, it detects DNA strand breaks and facilitates repair by recruiting proteins such as ATM, ATR, H2AX, and DNA-PK, which coordinate several DNA repair pathways like homologous recombination and non-homologous end joining. In cell death, excessive PARP1 activation can lead to ATP depletion and necrotic cell death, while controlled activity influences apoptosis. In inflammation, PARP1 regulates pro-inflammatory gene expression by modulating NF-κB signaling and cytokine production. In transcriptional regulation, it influences chromatin remodeling, RNA polymerase activity, and epigenetic modifications.
Through these roles, PARP1 maintains genomic stability, controls cell fate, and modulates immune responses, making it a key target in cancer and inflammatory disease therapies.
Resources
Are you looking for resources, click on the resource type to explore further.
Discover the versatility and precision of Homogeneous Time-Resolved Fluorescence (HTRF) technology. Our HTRF portfolio offers a...
Loading...


Recently viewed


How can we help you?
We are here to answer your questions.






























