

HTRF Human and Mouse Total CREBBP Detection Kit, 10,000 Assay Points


HTRF Human and Mouse Total CREBBP Detection Kit, 10,000 Assay Points






This HTRF kit enables the cell-based quantitative detection of Total CREBBP.
| Feature | Specification |
|---|---|
| Application | Cell Signaling |
| Sample Volume | 16 µL |
This HTRF kit enables the cell-based quantitative detection of Total CREBBP.



HTRF Human and Mouse Total CREBBP Detection Kit, 10,000 Assay Points



HTRF Human and Mouse Total CREBBP Detection Kit, 10,000 Assay Points



Product information
Overview
The HTRF Total CREBBP cell-based assay conveniently and accurately detects the levels of Total CREBBP within cells. This kit is compatible with the buffers from the Total p300 kit, so the same lysate can be used for analyses of both family members.
CREBBP (CREB Binding Protein, also known as CBP or KAT3A) and its paralog p300 (also called EP300 or KAT3B) are key transcriptional co-activators. These Lysine Acetyl Transferases (KATs) form the two-member KAT3 family. They control gene transcription by facilitating the assembly of transcriptional machinery and by acetylating specific lysine residues on core histones and on transcription coregulators. CREBBP and p300 act as network "hubs", with more than 400 proteins to regulate the expression of genes controlling several basic cellular processes, such as proliferation and homeostasis.
Mutations or chromosomal translocations involving CREBBP/p300 lead to gene dysregulation and can result in a variety of human diseases such as cancer, neurodegeneration, and inflammatory diseases. CREBBP/p300 PROTAC degraders have recently been developed for the treatment of human cancers.
Specifications
| Application |
Cell Signaling
|
|---|---|
| Brand |
HTRF
|
| Detection Modality |
HTRF
|
| Lysis Buffer Compatibility |
Lysis Buffer 1
Lysis Buffer 2
Lysis Buffer 3
Lysis Buffer 4
|
| Molecular Modification |
Total
|
| Product Group |
Kit
|
| Sample Volume |
16 µL
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Target Class |
Phosphoproteins
|
| Target Species |
Human
Mouse
|
| Technology |
TR-FRET
|
| Unit Size |
10,000 assay points
|
Video gallery

HTRF Human and Mouse Total CREBBP Detection Kit, 10,000 Assay Points

HTRF Human and Mouse Total CREBBP Detection Kit, 10,000 Assay Points

How it works
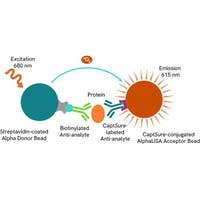
Total CREBBP assay principle
The HTRF Total CREBBP assay quantifies the expression level of CREBBP in cell lysate. Unlike Western Blot, the assay is entirely plate-based, and does not require gels, electrophoresis, or transfer. The Total CREBBP assay uses two labeled antibodies, one coupled to a donor fluorophore, the other to an acceptor. Both antibodies are highly specific for a distinct epitope on the protein. In presence of CREBBP in a cell extract, the addition of these conjugates brings the donor fluorophore into close proximity with the acceptor, and thereby generates a FRET signal. Its intensity is directly proportional to the concentration of the protein present in the sample, and provides a means of assessing the protein’s expression under a no-wash assay format.

Total CREBBP two-plate assay protocol
The 2-plate protocol involves culturing cells in a 96-well plate before lysis, then transferring lysates into a 384-well low volume detection plate before the addition of the Total CREBBP HTRF detection reagents. This protocol enables the cells' viability and confluence to be monitored.

Total CREBBP one-plate assay protocol
Detection of total CREBBP with HTRF reagents can be performed in a single plate used for culturing, stimulation, and lysis. No washing steps are required. This HTS-designed protocol enables miniaturization while maintaining robust HTRF quality.

Assay validation
Characterization of the mechanism of action of 3 CREBBP/p300 PROTACs
The experiments were carried out using the two-plate assay protocol for adherent cells. HeLa cells were plated in 96-well culture plates (70,000 cells/well in complete culture medium) and cultured overnight at 37°C-5% CO2. The day after, the cells were pre-treated for 1 hour with 1 µM Epoxomicin and treated during an additional 5h-incubation with 3 doses of each CREBBP/p300 PROTAC degrader (PROTAC CBP/p300 Degrader-1, Thalidomide-NH-CBP/P300 ligand 2, or dCBP-1), as well as with the warhead GNE-781 used as negative control (the same for the three PROTACs). The cells were lysed with 50 µL of supplemented lysis buffer #1, and 16 µL of lysate were then transferred into a low volume white microplate (ProxiPlate-384 Plus, Cat # 6008280/9) before the addition of 4 µL of the HTRF Total CREBBP or Total p300 kit detection antibodies (HTRF Total p300 kit, Cat # 64P300TPEG/Y). For both assays, the HTRF signal was recorded after incubating overnight. Cell viability was also assessed by transferring 5 µL of the same lysate into an HTRF 96-well low volume white plate (Cat # 66PL96005/025/100) and then adding 25 µL of ATPlite 1step detection reagent (ATPlite 1step Luminescence Assay System, Cat # 6016736/1/9). The luminescence signal was measured after a 10-min incubation in the dark.
After 5 hours of treatment, the three CREBBP/p300 PROTAC degraders induced a dose dependent signal decrease in both CREBBP and p300, whereas the warhead GNE-781 caused no change. This was as expected. As indicated on the ATPlite graph, no cytotoxic effect was observed in presence of the compounds. Finally, HTRF signals were recovered in presence of the proteasome inhibitor Expoxomicin. Taken together, our results unambiguously demonstrated PROTAC-induced CREBBP/p300 degradation which is mediated by the ubiquitin proteasome system.



Dose-effect and time-course studies of the 3 CREBBP/p300 PROTACs
The experiments were conducted on HeLa cells as mentioned in the previous section, except that the cells were treated with increasing concentrations of each CREBBP/p300 PROTAC (or warhead GNE-781) for 4 hours or 20 hours. As described before, Total CREBBP and Total p300 were detected using the corresponding HTRF kits, and cell viability was measured using the ATPlite 1step luminescence kit.
The three PROTACs induced a dose-dependent degradation of both CREBBP and p300, while the warhead GNE-781 did not. The compounds displayed comparable behavior towards the two proteins (e.g. potency and kinetics). The ranking of the three degraders was the same after the two treatment times, with Thalidomide-NH-CBP/P300 ligand 2 being 4 times and 10 times more potent than PROTAC CBP/p300 Degrader-1 and dCBP-1 respectively (based on DC50 values). The potencies of PROTAC CBP/p300 Degrader-1 and dCBP-1 were greatly improved after a 20h-treatment (from a factor 2.5 to 5), as well as the degradation efficacy of Thalidomide-NH-CBP/P300 ligand 2, which increased from 65% to 80%. It should be noted that the compounds did not induce noticeable cytotoxic effects, even after a 20h-treatment (ATPlite 1step assay, data not shown).




Validation of the specificity of Total CREBBP and Total p300 assays
HeLa cells were plated in 96-well culture plates (50,000 cells/well for Total CREBBP and 12,500 cells/well for Total p300) in complete culture medium and cultured overnight at 37°C, 5% CO2. The day after, the cells were transfected with ON-TARGETplus SMARTPool siRNAs specifically targeting human CREBBP or its paralog human p300, as well as with a non-targeting siRNA used as a negative control. Following a 24h-incubation with siRNAs, the medium was renewed for an additional 24h-incubation, and the cells were lysed with 50 µL of supplemented lysis buffer #1. After cell lysis, Total CREBBP and Total p300 were detected using the corresponding HTRF kits, and cell viability was measured using the ATPlite 1step luminescence kit, as described in the first section.
The siRNA experiments show that both HTRF Total CREBBP and Total p300 kits specifically measure the protein of interest and do not recognize the other family member. The treatment with the CREBBP siRNA induced a 70% signal decrease for the Total CREBBP assay, whereas the knockdown of p300 did not lead to any signal decrease in this assay. Reciprocally, the incubation with the p300 siRNA induced a 86% signal decrease for the Total p300 assay, while the silencing of the CREBBP gene did not lead to any signal decrease in this assay. The ATPlite luminescence signal remained quite stable in presence of the CREBBP and p300 siRNAs, demonstrating that the HTRF signal decrease is unrelated to cytotoxic effects.


Assessment of total CREBBP levels in various human and mouse cell lines
The experiments were carried out using the two-plate assay protocol for adherent cells. The human cell lines SH-SY5Y, MCF7, HeLa, A431, and A549, as well as the mouse cell lines NIH-3T3 and Neuro-2a, were plated in 96-well culture plates (100,000 cells/well in complete culture medium) and cultured at 37°C, 5% CO2 for 24 hours. The day after, the cells were lysed with 50 µL of supplemented lysis buffer #1, and 16 µL of lysate were transferred into a low volume white microplate (ProxiPlate-384 Plus, Cat # 6008280/9) before the addition of 4 µL of HTRF Total CREBBP kit detection antibodies. The HTRF signal was recorded after an overnight incubation.
The HTRF Total CREBBP kit is compatible with human and mouse cell models and efficiently detects CREBBP in cell lines containing variable expression levels of the protein.

HTRF Total CREBBP assay compared to Western Blot
HeLa cells were seeded in a T175 flask and cultured at 37°C, 5% CO2 in complete culture medium until ~ 90% confluency was reached. The cells were lysed with 3 mL of supplemented lysis buffer #1 and serial dilutions of the lysate were performed using supplemented lysis buffer. To detect Total CREBBP, 16 µL of each dilution were transferred into a low volume white microplate (ProxiPlate-384 Plus, Cat # 6008280/9) and 4 µL of HTRF Total CREBBP detection reagents were added. Equal amounts of lysates were loaded on a gel for a side-by-side comparison with Western Blot.
Using the HTRF Total CREBBP assay, 500 cells/well were enough to detect a significant signal, while 4,000 cells were needed to obtain a minimal chemiluminescent signal using Western Blot. Therefore, in these conditions the HTRF Total CREBBP assay was 8 times more sensitive than the Western Blot technique.

Simplified pathway
CREBBP/p300 Signaling Pathway
The transcriptional co-activators CREBBP and p300 regulate gene expression by facilitating the assembly of transcriptional machinery and by acetylating core histones and other transcriptional regulators such as the transcription factors (TFs) c-Myc, p53 and FoxO1.
CREBBP and p300 are central to many important signaling pathways activated by intracellular or extracellular signals. They regulate growth factors and hormones signaling such as the one mediated by the nuclear hormone receptors AR (Androgen Receptor) and ER (Estrogen Receptor). Other examples of signals are hypoxia, cellular stress (involving the MAP kinases p38, JNK and ERK), high glucose, GPCR agonists (inducing cAMP signaling) and cytokines (mediated by the JAK/STAT pathway). The TLR/IRF-3 pathway is also regulated by CREBBP/p300 to control innate immune responses. Both CREBBP and p300 are required for histone acetylation at BRD4-occupied sites (e.g., H3K27ac) and for BRD4 binding at gene promoters and enhancers to regulate associated gene transcription.
All these pathways ultimately use CREBBP and p300 as downstream effectors in the nucleus to control key cellular functions such as cell cycle control and differentiation, embryonic development, and homeostasis.

Resources
Are you looking for resources, click on the resource type to explore further.
This guide provides you an overview of HTRF applications in several therapeutic areas.
Loading...


Recently viewed
How can we help you?
We are here to answer your questions.






























