
NEXTFLEX Small RNA Sequencing Kit V4


NEXTFLEX Small RNA Sequencing Kit V4






| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | Small RNA-seq |









Supporting products you might need
Are you working with blood samples?
Product information
Overview
The NEXTFLEX small RNA sequencing kit v4 uses patent-pending technology to provide a completely gel-free small RNA library preparation solution for Illumina and Element sequencing platforms.
- Completely gel-free protocol from purified miRNA or as little as 1 ng total RNA, no PAGE cleanup required
- Automation-ready on Sciclone™ G3 NGSx, Sciclone G3 NGSx iQ™, and Zephyr™ G3 NGS workstations for walk-away scalability
- Low-bias ligation plus dimer reduction chemistry improve data quality from challenging biofluids (plasma, serum, urine, CSF) and extracellular vesicles.
- Internal benchmarking shows up to a 45% increase in unique miRNA species detected from as little as 1 ng of total RNA, supporting robust differential-expression analysis and novel small RNA discovery
- Libraries ready in ~ 6 hours, supporting rapid biomarker and functional genomics studies
- Includes 384 Unique Dual Index (UDI) barcodes for high-throughput multiplexing and minimal index hopping on Illumina®/Element® platforms
- Quality assured through rigorous testing of every lot
Explore our Dharmacon™ miRNA modulation reagents to enhance or inhibit miRNA levels as part of your functional studies.
Additional product information
- Ready-to-run Sciclone/Zephyr automation scripts with deck maps, tip-usage estimates, and 96-/384-well timing
- No PAGE excision – standard magnetic bead clean-ups eliminate laborious gel slices while preserving low-input yield
- Wide sample compatibility: plasma, serum, urine, CSF, exosomal RNA, etc
- Captures diverse < 200 nt species – miRNA, siRNA, piRNA, tRNA-derived fragments in a single ligation-based workflow
- Scalable study design – one workflow supports pilot studies (8 samples) through 384-plex UDI projects without protocol changes
We compared different vendors, and we found that the small RNA sequencing kit from Revvity was the one that performed best.
- Dr. Sören Franzenburg, Head of NGS platform, Kiel University
Gel-free small RNA library prep with reduced bias
The NEXTFLEX Small RNA-Seq Kit v4 offers a completely gel-free library prep workflow for Illumina® and Element® sequencers. Optimized 3′ and 5′ adapter-ligation chemistry, together with proprietary dimer-reduction oligonucleotides, minimizes adapter-dimer formation and sequence-dependent ligation bias without the need for PAGE cleanup.
The six-hour protocol reliably produces high-complexity libraries from as little as 1 ng of total RNA derived from plasma, serum, urine, CSF, cultured cells or tissue lysates. Internal benchmarking shows higher on-target mapping rates and a larger repertoire of unique miRNA species than standard small RNA workflows.
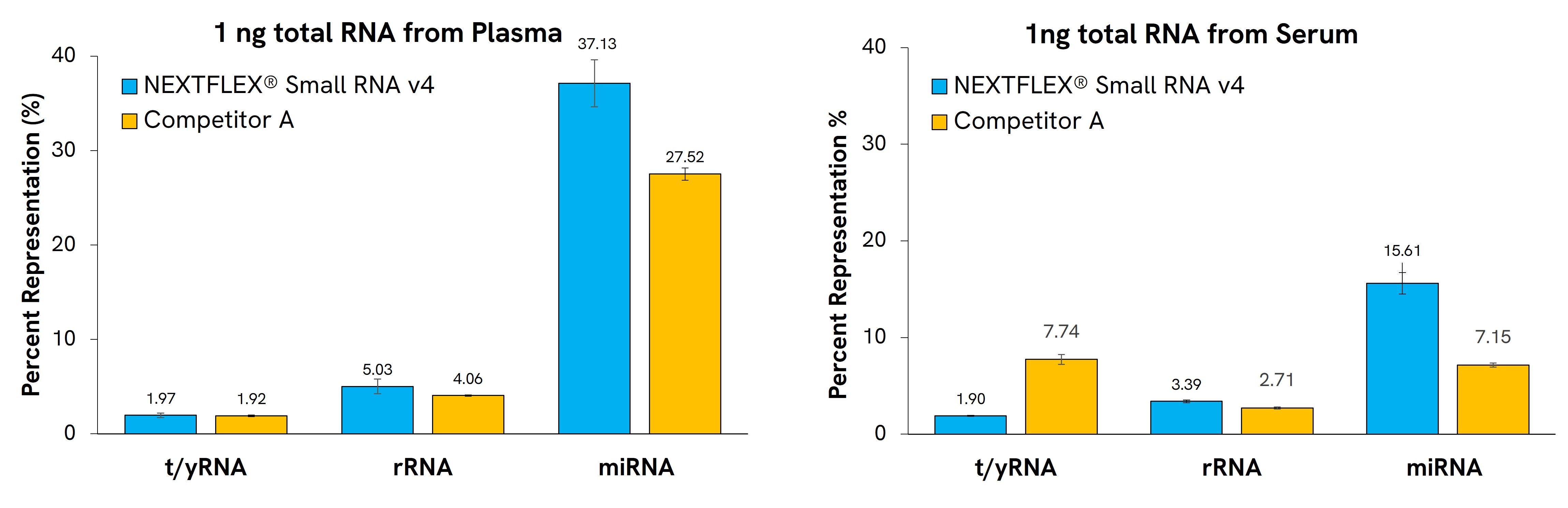
To further enhance sensitivity in biofluid studies, the kit includes tRNA and YRNA blocking oligos, reducing over-represented fragments and enriching true miRNA reads (Figures 1 & 2).

Figure 1. NEXTFLEX Small RNA-Seq Kit v4 increases miRNA reads while reducing tRNA/YRNA carry-over. Libraries from 1 ng plasma RNA were sequenced and aligned to miRNA, tRNA, and YRNA reference sets. Compared with a competitor workflow, the NEXTFLEX Small RNA kit aligns with more miRNAs while, while minimizing reads associated with t/YRNA and RNA, improving effective sequencing depth for downstream small-RNA analysis.
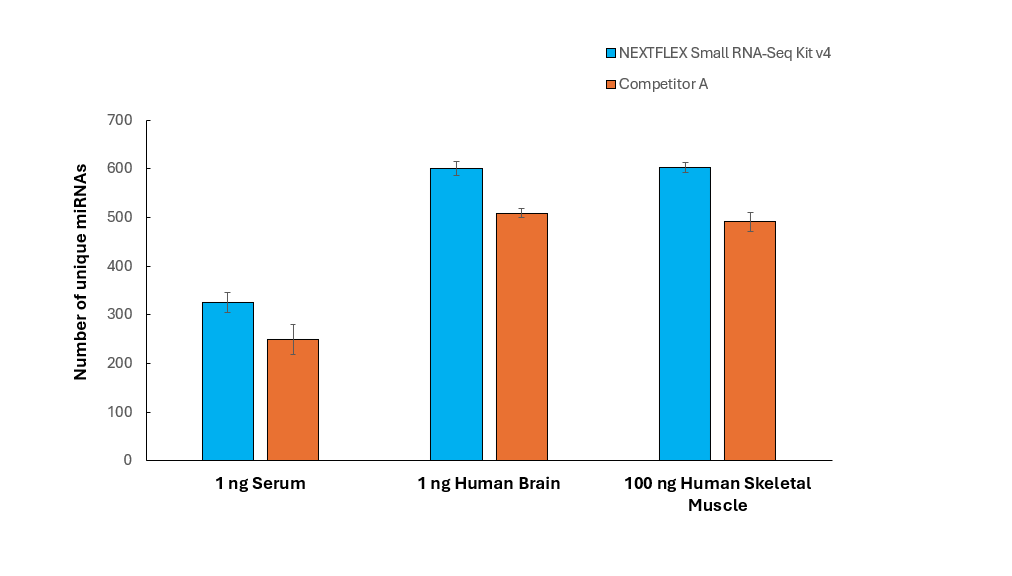
Figure 2: NEXTFLEX Small RNA-Seq Kit v4 detects higher number of unique miRNA species across different sample types
Exosomal RNA (exoRNA) Library Prep for Liquid Biopsy miRNA Profiling
The gel-free NEXTFLEX Small RNA-Seq Kit v4 workflow is equally effective for exosomal RNA (exoRNA). Even when starting with the picogram-level miRNA inputs typical of exosome preparations, the protocol generates libraries with high mature-miRNA alignment rates and negligible adapter-dimer carry-over (Figure 3). This enables reliable profiling of miRNAs derived from extracellular vesicles for liquid-biopsy biomarker discovery and other low-input applications.
Improved miRNA discovery
Internal benchmarking shows that the NEXTFLEX Small RNA-Seq Kit v4 recovers up to 45% more unique miRNA species than a competitor small RNA workflow when starting with as little as 1 ng of total RNA (Figure 4). This comprehensive profiling is essential for understanding the full spectrum of miRNA expression in each sample enabling a more complete view of post-transcriptional regulation and strengthens downstream differential-expression and biomarker-discovery analyses. The kit incorporates a highly processive rRT enzyme able to read through some of the common covalent modifications appearing in miRNA.
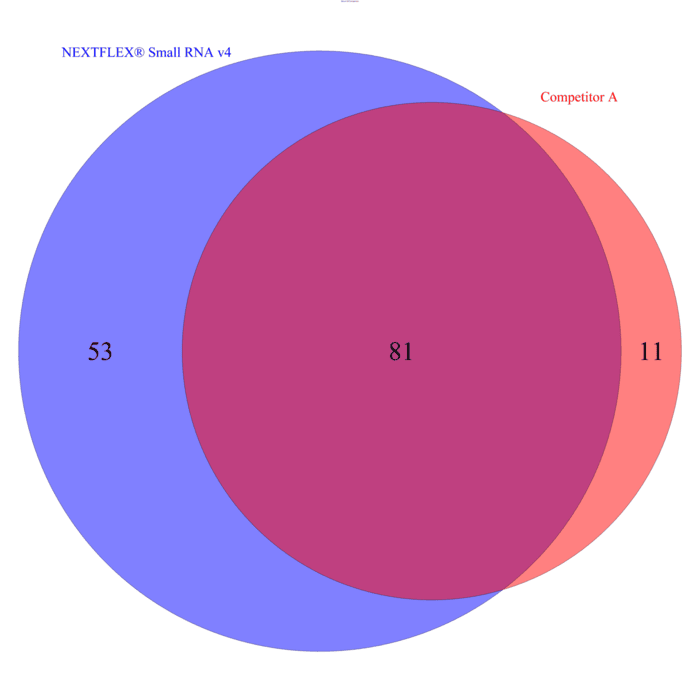
Figure 3. NEXTFLEX Small RNA v4 identifies 45% more unique miRNA species than competitor workflows. Venn diagram of libraries prepared from 1 ng serum RNA shows 81 miRNAs detected with both kits, an additional 53 miRNAs unique to NEXTFLEX, and 11 unique to the comparator.
Ribosome profiling (Ribo-Seq) and nascent-RNA transcriptomics
The gel-free NEXTFLEX Small RNA-Seq Kit v4 readily converts ribosome-protected fragments (28–30 nt RPFs) and nascent transcripts into sequencing libraries for Ribo-Seq and nascent transcriptomics application. By adapting small RNA library preparation methods, researchers can quantify translational efficiency, map ribosome-stall sites, and reveal upstream open-reading frames, critical insights for gene-expression control, disease-mechanism studies, and drug-target discovery.
“We frequently use Revvity small RNA sequencing kits for Ribosome- and Disome-profiling. It offers a very streamlined library generation and allows for easy customization to adapt to experimental needs. With this, it consistently delivers good results. Additionally, the technical support from Revvity’s team is excellent and very helpful which further enhanced our very positive experience with this Revvity product.”
- Dr. Tobias Schmidt, CRUK Scotland Institute
Automation & 384-Plex Multiplexing
Validated scripts make the NEXTFLEX workflow easy to setup on Sciclone G3 NGSx / iQ and Zephyr G3 workstation. It is also readily adapted to other liquid handlers. Up to 384 Unique Dual Index (UDI) barcodes are available, enabling high-throughput multiplexing on Illumina® and Element® sequencers while minimizing index hopping.
Maximize miRNA mapped reads with NEXTFLEX blood miRNA blockers
Boost usable miRNA reads in blood and plasma with NEXTFLEX Blood miRNA Blockers. These oligos selectively deplete highly abundant miR-486-5p, miR-92a-3p, and miR-451a—species that can consume 50–70% of reads so more sequencing depth is devoted to biologically informative miRNAs.
We have been using Revvity small RNA sequencing kits for a number of years, and we are satisfied with the results and the support received.
- Dr. Marcel van Herwijnen, Researcher/Lab Manager, Maastricht University
Boost Functional Insights with Dharmacon™ miRNA Modulators
Pair the NEXTFLEX® Small RNA-Seq Kit v4 with Dharmacon™ miRNA mimics and inhibitors to quickly elevate or silence target miRNAs, confirm their biological impact in any cell system, and move seamlessly from expression profiling to functional validation in a single streamlined workflow.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Barcodes |
1 - 8
|
| Product Group |
Small RNA-seq
|
| Shipping Conditions |
Dual Temperature
|
| Unit Size |
8 rxns
|
Video gallery
Citations
Profiling from extracellular vesicles
- Apostolov, A., Mladenović, D., Tilk, K. (2025) Multi-omics analysis of uterine fluid extracellular vesicles reveals a resemblance with endometrial tissue across the menstrual cycle: biological and translational insights, Human Reproduction Open, https://doi.org/10.1093/hropen/hoaf010.
- Marocco. F. (2025) The RNA binding protein PCBP2 is a regulator of miRNAs partition between cell and extracellular vesicles. [Doctoral dissertation, Sapienza University of Rome]. IRIS Uniroma1. https://iris.uniroma1.it/retrieve/6d5c8022-59e2-44ec-b8b6-eb7a6ea8cc6e/Tesi_dottorato_Marocco.pdf.
- Pan, Y., Meng, L., Yoshida, K., … & Asakawa, S. (2025). Comparative and functional analysis of exosomal microRNAs during semelparous reproduction in ayu fish. Journal of Extracellular Biology, 4(3), e70038. https://doi.org/10.1002/jex2.70038
- García-Concejo, A., Schwager, A., … & Blanco, B. (2025). Exosome-derived miRNAs distinguish septic shock from non-septic shock in postoperative patients. Journal of Translational Medicine, 23, XXX. (PMID: 40033446)
Biomarker discovery/validation
- Lill C. M., et al. (2025) EPIC4ND: European Prospective Investigation into Cancer and Nutrition follow-up for neurodegenerative diseases. medRxiv 2025.01.29.25321340; doi: https://doi.org/10.1101/2025.01.29.25321340.
- Chu, C. P., Nabity, M. B. (2025) Technical considerations and review of urinary microRNAs as biomarkers for chronic kidney disease in dogs and cats. Vet Clin Pathol. 2025; 00: 1-19. doi:10.1111/vcp.13412.
- Karere, G. M., Hsu, F.-C. Hepple, R. T. et al. (2025) MicroRNA signatures of VO2peak in older adult participants of the Study of Muscle, Mobility and Aging. bioRxiv 2025.01.08.631999; doi: https://doi.org/10.1101/2025.01.08.631999.
- Love, C. G., Coombs, L., & Van Laar, R. (2024). RNA-seq validation of microRNA expression signatures for precision melanoma diagnosis and prognostic stratification. BMC Medical Genomics, 17, 256. https://doi.org/10.1186/s12920-024-02028-w
Other
- Almeida, M.V., Blumer, M., Yuan, C.U. et al. Dynamic co-evolution of transposable elements and the piRNA pathway in African cichlid fishes. Genome Biol 26, 14 (2025). https://doi.org/10.1186/s13059-025-03475-z.
- Rossner, p., Javorkova, E., Sima, M. et al. Skin wound healing: the impact of treatment with antimicrobial nanoparticles and mesenchymal stem cells. (2025) Authorea. February 17, 2025. DOI: 10.22541/au.173977945.54128573/v1.
- Tahiri, I., Llana, S.R., Díaz-Castro, F. et al. AgRP neurons shape the sperm small RNA payload. Sci Rep 15, 7206 (2025). https://doi.org/10.1038/s41598-025-91391-4.
Ribo-Seq
- Ting MKY, Gao Y, Barahimipour R, et al. Optimization of ribosome profiling in plants including structural analysis of rRNA fragments. Plant Methods. 2024;20(1):143. doi: 10.1186/s13007-024-01267-3.
- Bohlen J, Fenzl K, Kramer G, Bukau B, Teleman AA. Selective 40S footprinting reveals cap-tethered ribosome scanning in human cells. Molecular Cell. 2020;79(4):561-574.e5. doi: 10.1016/j.molcel.2020.06.005.
- Hoerth K, Reitter S, Schott J. Normalized Ribo-Seq for quantifying absolute global and specific changes in translation. Bio-Protocol. 2022;12(4):e4323. doi: 10.21769/BioProtoc.4323
FAQs
-
What types of RNAs are included in the Small RNA library?
-
Do you have any recommendations for extraction of RNA for Small RNA sequencing?
-
Are the libraries produced by NEXTFLEX Small RNA Sequencing Kit stranded/directional?
-
Can I use total RNA, or do I have to isolate or enrich the sample for small RNA?
-
What happens if the quality of my RNA is very low?
-
How should my NEXTFLEX Small RNA libraries be trimmed?
-
What should I do if my Small RNA may not have 5' Phosphate and 3' OH groups?
-
How are the barcodes introduced in the NEXTFLEX Small RNA libraries?
-
What are the recommended read length and settings for sequencing?
-
What is the number of reads required for small RNA sequencing?
-
Do I have to use custom Illumina® sequencing primers for the libraries generated with NEXTFLEX Small RNA Sequencing Kit v4?
-
What do I need to do to run the NEXTFLEX Small RNA libraries on the AVITI™ sequencer from Element® Biosciences?
-
Are the Monarch™ Spin RNA Cleanup Kits (NEB) compatible with NEXTFLEX reagents for RNA library prep?
-
Do you have recommendations for analysis of small RNA sequencing data?
-
Do you have recommendations on EV isolation compatible with downstream Small RNA sequencing?
Resources
Are you looking for resources, click on the resource type to explore further.
Extracellular vesicles (EVs) represent a fascinating frontier in biomarker discovery. Secreted by virtually all cell types, these...
This flyer describes the benefits of the NEXTFLEX® Small RNA-Seq Kit v4 kit
In this application note we describe a simplified, commercially available protocol encompassing exoRNA extraction and preparation...
This excel file includes the sequences of the indexes for the UDI primers included in the NEXTFLEX Small RNA-seq kit v4.
NEXTFLEX® Small RNA-Seq Kit v4 has a streamlined, gel-free workflow delivering exceptional miRNA mapping and discovery rates even...
This poster illustrates the effieciency of the NEXTFLEX® Small RNA-Seq Kit v4 kit of discovering biomarkers.
SDS, COAs, manuals and more
Are you looking for technical documents related to the product? We have categorized them in dedicated sections below. Explore now or request your COA/TDS, SDS, or IFU/manual.
- LanguageEnglishCountryUnited States
- Resource TypeManualLanguageEnglishCountry-
- Resource TypeManualLanguageEnglishCountry-
- Resource TypeManualLanguageEnglishCountry-
Loading...


How can we help you?
We are here to answer your questions.
































