Barcoded NGS Adapters and Blockers
All of Revvity’s NGS adapters incorporate unique dual indexes (UDI) and are compatible with Illumina, Watchmaker, Twist, and any other TruSeq style library prep kit. We also offer adapters with both unique molecular adapters (UMI) and UDIs. The information below will help you decide what adapter is best for your application.
Blockers are essential in hybridization enrichment workflows to reduce off-target effects and optimize the performance of the hybridization process. Our blockers are designed to bind to Illumina adapter sequences, which are the most common sequences present in the hybridization reaction. This ensures that the probes will bind preferentially bind to the intended target sequences.
For research use only. Not for use in diagnostic procedures.
Unique Dual Indexes (UDI)
UDIs (represented as i5 and i7 in Figure 1) are added to both ends of an NGS library molecule. This enables researchers to multiplex multiple samples in parallel. We offer sets of 384 adapters (8 nt index) and 1536 adapters (10 nt index).
The presence of dual indices minimizes index hopping—a phenomenon where a read is incorrectly assigned to the wrong index due to sequencing errors.
“Revvity has a set of 1536 validated UDI adapters that we have been regularly using for 5 years. Using the Illumina® NovaSeq® X Plus with this UDI set from Revvity we can sequencing 12,288 samples across 8 lanes of the 10B or 25B flow cells. This dramatically lowers our sequencing cost per sample!”
- Dr. Charlie Johnson, Director of Texas A&M Agri Life Genomics and Bioinformatics Service
Unique Molecular Identifier (UMI): A single 9-base-pair UMI is incorporated into each library. Revvity’s UMIs provide over 250,000 unique combinations to tag individual molecules within a sample library. UMIs are relevant in applications where we want to differentiate PCR duplicates (which share the same UMI sequence) from true copy numbers (each copy having a distinct UMI sequence). They are also used to distinguish true variants from errors introduced during library preparation.
Advantages of Revvity’s Ligation-Based Adapters:
Incorporating full-length UDI and UMI sequences via ligation (instead of PCR) reduces the risk of introducing errors. This approach allows researchers to leverage these features effectively in PCR-free workflows. We recommend the use of the NEXTFLEX UDI-UMI barcodes to improve variant calling, gene expression analysis, heterogeneous tumor sample sequencing, ctDNA sequencing, deep exome sequencing, single-cell RNA-seq, and haplotyping via linked reads.
- Every lot is functionally verified and tested for index purity by sequencing.
Selection Guide:
| Config | Index Length | Multiplexing options | Sequencers | Library Type Compatibility | Kit |
|---|---|---|---|---|---|
| UDI | 8 | 384 samples | Illumina® & Element® platforms* | TruSeq style with ligation-based adapters |
NEXTFLEX® Unique Dual Index Barcodes
|
| 10 | 1536 samples |
NEXTFLEX® UDI Barcodes (Set of 1536)
|
|||
| UDI-UMI | 10 | 96 samples** | NEXTFLEX® UDI-UMI Barcodes |
*When utilized with Aviti Cloudbreak™ Freestyle chemistry
**Contact us for availability beyond 96 UDI-UMIs.
Figure 1. Example of NEXTFLEX® Adapter design

Figure Insert. DNA or RNA fragment from a sample. P5 and P7. Flow cell binding sites for Illumina® platforms, SP1 and SP2. Binding sites for sequencing primers, i5 and i7. Short sequences (8 or 10 bp) used to identify a particular sample (UDI). UMI. 9 bp sequence used to uniquely tag each molecule within a library.
Universal Blockers
During target enrichment, adapters can interact with the complementary adapter sequence strand of another library molecule. The NEXTFLEX® Universal Blockers are designed to prevent this concatemerization of library molecules. By blocking this complementary Illumina adapter sequence interaction, the number of on-target reads improves dramatically and the depth of enrichment increases. They are not affected by index length or the presence of a unique molecular index (UMI). The NEXTFLEX Universal Blockers are compatible with ligation-based (TruSeq style) and tagmentation-based (Nextera style) library prep kits such as those from IDT and Twist, for Illumina® and Element® sequencing platforms
Figure 2. NEXTFLEX® Universal Blocker design
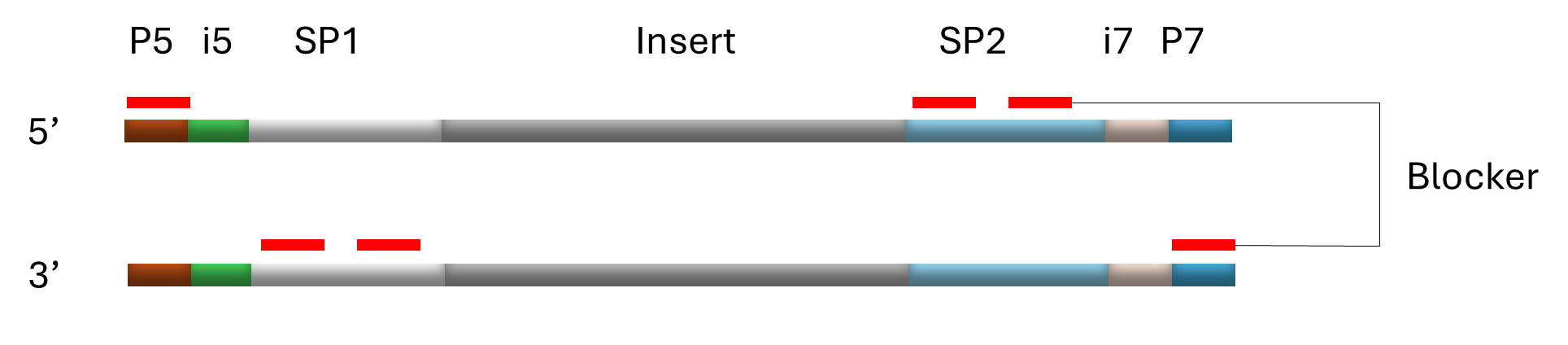
Figure Insert. The NEXTFLEX® Universal Blockers are composed of a proprietary mixture of modified oligos biding to P5, P7, SP1 and SP2 sites.