Customer experiences
"We are using model chemagic™ 360 (Automated Nucleic Acid Extraction system) installed in our laboratory for COVID-19 testing. We are running 32 min. short protocol (recently updated by the chemagen Team) and getting good result with our RT PCR. Due to this short protocol we are able to run approx. 4000 samples / day with two instruments, The instrument supplied has been functioning well, satisfying the requirements of our applications and meeting our expectations for quality and reliability of the data generated." Department of Microbiology, Government of Rajasthan, S.M.S. Medical School, Jaipur"
"We are using model chemagic™ 360 (Automated Nucleic Acid Extraction system) installed in our laboratory for COVID-19 testing. We are running 32 min. short protocol (recently updated by the chemagen Team) and getting good result with our RT PCR. Due to this short protocol we are able to run approx. 4000 samples / day with two instruments, The instrument supplied has been functioning well, satisfying the requirements of our applications and meeting our expectations for quality and reliability of the data generated." Department of Microbiology, Government of Rajasthan, S.M.S. Medical School, Jaipur"
"We had good earlier experience with the chemagic 360 nucleic acid extraction system and found it to be highly versatile, filling many existing and future needs in our operations. When the viral extraction kit and RT-PCR kit became available it was a natural choice to adopt the complete workflow from a single, reliable and local supplier."
Biopsense, Jyväskylä, Finland
SARS-CoV-2 RNA detection
With an ultrafast run-time of 18 min, SARS-CoV-2 RNA extraction is performed from plasma, swabs or saliva in the high-throughput workflow on the chemagic 360 instrument. Please find below an interesting example of use.
With an ultrafast run-time of 18 min, SARS-CoV-2 RNA extraction is performed from plasma, swabs or saliva in the high-throughput workflow on the chemagic 360 instrument. Please find below an interesting example of use.
PCR detection of mcroorganisms
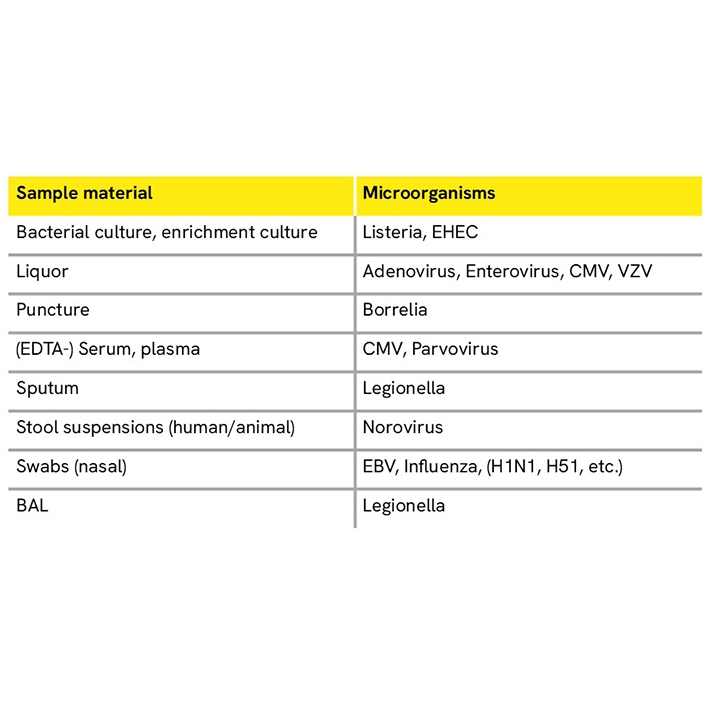
Detect diverse microorganisms from a wide range of sample material including stool, whole ticks, and body fluids using the chemagic Viral NA/gDNA 200 Kit H96 (CMG-1049-A) for low volumes and the chemagic Body Fluid 1k Kit H96 (CMG-1131) for high volumes. The chemagic Viral DNA/RNA 300 Kit H96 (CMG-1033-S) can be employed for clear body fluids such as plasma, serum, swabs, and saliva.
Detect diverse microorganisms from a wide range of sample material including stool, whole ticks, and body fluids using the chemagic Viral NA/gDNA 200 Kit H96 (CMG-1049-A) for low volumes and the chemagic Body Fluid 1k Kit H96 (CMG-1131) for high volumes. The chemagic Viral DNA/RNA 300 Kit H96 (CMG-1033-S) can be employed for clear body fluids such as plasma, serum, swabs, and saliva.
Compatible with third party PCR providers (including the EURORealTime™ PCR Systems from EUROIMMUN), these workflows are regularly employed in demanding, high throughput workflows in research and diagnostic labs around the world.
Rapid Candida auris detection with real-time PCR
Candida auris genetic testing is crucial for identifying and tracking the spread of this multidrug-resistant fungal pathogen, aiding in infection control measures and guiding appropriate antifungal treatment strategies.
Candida auris genetic testing is crucial for identifying and tracking the spread of this multidrug-resistant fungal pathogen, aiding in infection control measures and guiding appropriate antifungal treatment strategies.
Revvity has developed a C. auris detection real-time PCR workflow that uses the chemagic™ Pathogen NA Kit H96 (CMG-1033-G) on the chemagic 360 instrument for high throughput nucleic acid extraction from skin swabs, environmental surface swabs, or laboratory cultures.
SARS-CoV-2 wastewater surveillance
Wastewater testing for SARS-CoV-2 RNA can provide real-time surveillance of viral spread at the community level. Due to large sample volumes, sample inhomogeneity and continuous flow, special pre-treatment and concentration of samples are often required e.g. with the use of filters11 or by direct extraction12.
Wastewater testing for SARS-CoV-2 RNA can provide real-time surveillance of viral spread at the community level. Due to large sample volumes, sample inhomogeneity and continuous flow, special pre-treatment and concentration of samples are often required e.g. with the use of filters11 or by direct extraction12.
Find more details in the app note.

Cost-effective multiplex testing of tick-borne pathogens
Tick-borne pathogens are difficult to detect due to poor sensitivity and the lack of comprehensive tick-borne panels to cover pathogens endemic to different regions.
Tick-borne pathogens are difficult to detect due to poor sensitivity and the lack of comprehensive tick-borne panels to cover pathogens endemic to different regions.
With the chemagic Viral NA/gDNA 200 Kit H96 (CMG-1049-A), extractions can be performed from blood, synovial fluid or whole ticks.
References:
- MacKay, M.J., Hooker, A.C., Afshinnekoo, E. et al. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat Biotechnol (2020).
- Ludwig, K.U. et al. LAMP-Seq enables sensitive, multiplexed COVID-19 diagnostics using molecular barcoding. Nat Biotech (2021).
- Siedner, M.J., Moorhouse, M.A., Simmons, B. et al. Reduced efficacy of HIV-1 integrase inhibitors in patients with drug resistance mutations in reverse transcriptase. Nat Commun (2020).
- Kleines, M., Schellenberg, K., Ritter, K. Efficient Extraction of Viral DNA and Viral RNA by the Chemagic Viral DNA/RNA Kit Allows Sensitive Detection of Cytomegalovirus, Hepatitis B Virus, and Hepatitis G Virus by PCR. J. Clin. Micribiol. (2003).
- Bliss, J. et al. High Prevalence of Shigella or Enteroinvasive Escherichia coli Carriage among Residents of an Internally Displaced Persons Camp in South Sudan. Am. J. Trop. Med. Hyg. (2018).
- Michel J., Targosz A., Rinner T., Bourquain D., Brinkmann A., Sacks JA., Schaade L., Nitsche A.. Evaluation of 11 commercially available PCR kits for the detection of monkeypox virus DNA. (2022)
- Tang, Y. et al. An Isothermal, Multiplex Amplification Assay for Detection and Genotyping of Human Papillomaviruses in Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagnost. (2020).
- Rajeevan, M.S. et al. NanoString Technology for Human Papillomavirus Typing. Viruses. (2021).
- Lefèvre, A.C. et al. The Clinical Value of Measuring Circulating HPV DNA during Chemo-Radiotherapy in Squamous Cell Carcinoma of the Anus. Cancers. (2021).
- LaTurner, Z. W . Zong, D. M., Kalvapalle, P., Gamas, K. R., Terwilliger, A., Crosby, T., Ali, P., Avadhanula, V., Santos, H. H., Weesner, K., Hopkins, L., Piedra, P. A., Maresso, A. W., & Stadler, L. B.(2021). Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Research, 197. (2021).
- West, N. W., Vasquez, A. A., Bahmani, A., Khan, M. F., Hartrick, J., Turner, C. L., Shuster, W., & Ram, J. L. Sensitive detection of SARS-CoV-2 molecular markers in urban community sewersheds using automated viral RNA purification and digital droplet PCR. The Science of the Total Environment, 847, 157547. (2022).


Filters
1 - 4 of 4 Products and Services
The chemagic™ 360 instrument is a flexible compact solution for automated nucleic extraction from diverse sample materials.
The chemagic™ Prime™ Instrument is a fully automated solution offering hands-free sample transfer, DNA and RNA isolation, normalization (optional), and PCR setup for research applications. This validated, single supplier solution offers high quality DNA and RNA which can be extracted from a variety of matrices.
The chemagic™ Prepito® instrument is an automated system for high-quality DNA/RNA isolation based on proven M-PVA Magnetic Bead Technology.
The chemagic™ Viral NA/gDNA 200 Kit H96 can be used for automated viral nucleic acid purification from up to 200 µl blood and body fluids together with chemagic 360 instrument equipped with the chemagic 96 Rod Head.