Overview
This information is relevant for LANCE™ Ultra kinase assays that use a ULight™-labeled substrate (2-component assay). All concentrations listed represent the final concentration in the reaction; all assays in triplicate:
1. Substrate identification/selection (if applicable)
- Kinase reaction (10 µL)
- For Ser/Thr kinases, use 20 nM kinase
- For Tyr kinases, use 1 nM or less kinase (a low concentration will likely need to be used, due to possible generic anti-phosphotyrosine antibody depletion).
- 50 or 100 nM ULight-substrate (refer to the tech data sheet for the product to see what concentration to use)
- 100 µM ATP
- Control: no ATP
- Incubate 60 minutes
-
Detection (brings volume to 20 µL)
- 10 mM EDTA
- 2 nM Europium antibody

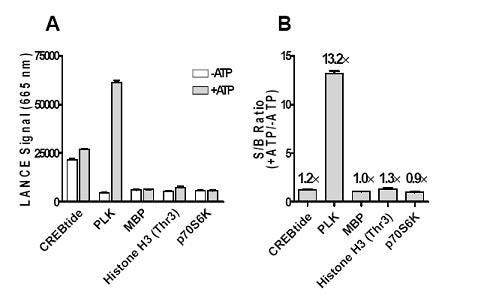
Figure 1. Figure (from Ser/Thr KinaSelect™ manual). Selection of the optimal substrate for the Aurora A kinase. The Aurora A kinase (Carna Biosciences) at 20 nM was incubated with either ULight-CREBtide (Ser133), ULight-PLK (Ser137), ULight-MBP, ULight-Histone H3 (Thr3) or ULight p70 S6K (Thr389) peptide in the presence of 200 µM ATP. Kinase reactions were terminated after 1 hour by the addition of EDTA followed by the addition of 1X LANCE Detection buffer containing the corresponding Eu-labeled antibody at a final concentration of 2 nM. Signal was read after 1 hour. A) LANCE signal at 665 nm. B) S/B ratio: +ATP/-
2. Enzyme titration vs. time-course kinetics
- Choose 4-6 concentrations of kinase
- For Ser/Thr kinases, appropriate kinase concentrations will generally range from 10 pM to 20 nM (final concentration in 10 µL kinase reaction)
-
For Tyr kinases, choose concentrations near and below the concentration listed in the Tyrosine kinase substrate selection guide. In general, your range will be below 1 nM (final concentration in 10 µL kinase reaction). See below for generic anti-phosphotyrosine antibody depletion.
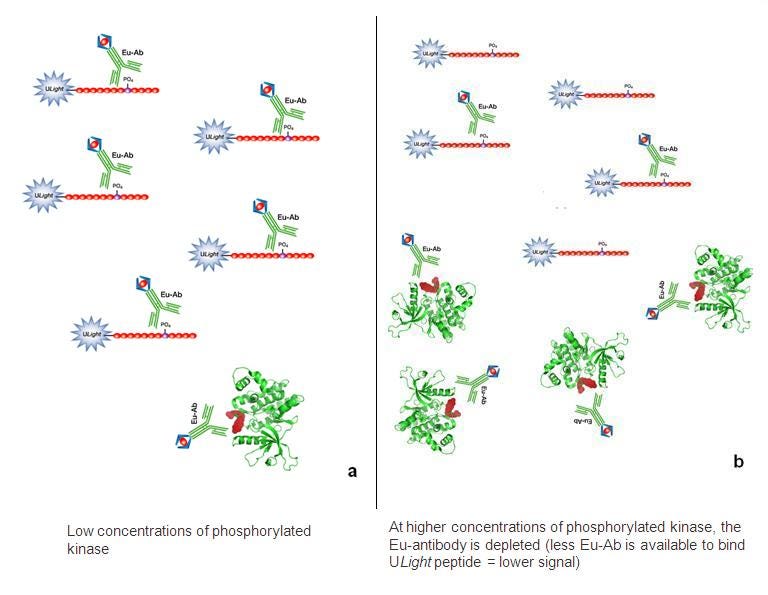
- Run time course experiment at each concentration of kinase (0, 5 min, 15 min, 30 min, 60 min, and 90 min). This is the time course for the kinase reaction step. You will stop the kinase reaction by adding EDTA. For the 0 min time point, add kinase + substrate + EDTA, incubate 1 minute, then add ATP (only after the EDTA has been added).
- 100 µM ATP; 50 or 100 nM ULight-substrate (refer to the tech data sheet for the product to see what concentration to use), 2 nM Eu-anti-phospho-antibody.
-
Control: 0 nM kinase

Figure on left (from Tech note): Aurora B enzyme was incubated at concentrations ranging from 10 to 250 pM with 50 nM ULight-Histone H3 peptide and 20 µM ATP. Kinase reactions were terminated after 0 to 120 minutes by the addition of EDTA. The anti-phospho-Histone H3 (Ser10) antibody was used. Figure on the right: mTOR enzyme was incubated at concentrations ranging from 1.25 to 20 nM with 50 nM ULight-p70 S6K peptide and 200 µM ATP. Kinase reactions were terminated after 0 to 120 minutes by the addition of EDTA.
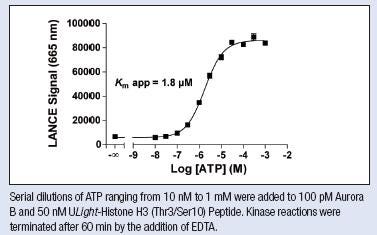
3. ATP titration
- ATP concentrations: 300 µM, 100 µM, 30 µM, 10 µM, 3 µM, 1 µM, 300 nM, 100 nM, 30 nM, 10 nM, and 0 nM
- Choose a kinase concentration and time point from your previous data. You will want to pick a concentration and time point in the linear range of your graph that give good S:B while conserving kinase
- 50 or 100 nM ULight-substrate (refer to the tech data sheet for the product to see what concentration to use), 2 nM Europium antibody
-
Determine Km for ATP
4. Inhibition curve
- ATP concentration at or below the Km determined above
- Same kinase concentration and time point used in your ATP titration experiment
- Inhibitor concentrations as relevant
-
50 or 100 nM ULight-substrate (refer to the tech data sheet for the product to see what concentration to use), 2 nM Europium antibody
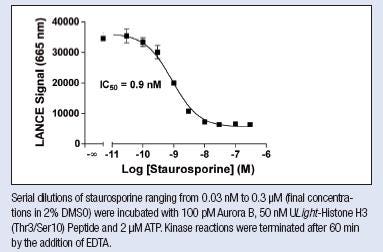
Figure from Tech note
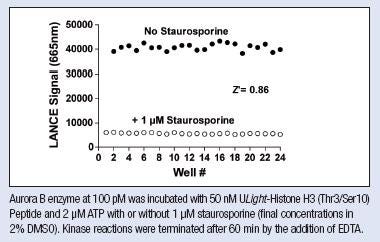
5. Z' factor determination
In a 384-well OptiPlate™, set up one row with kinase + substrate + ATP as optimized above + inhibitor (or exclude ATP). Set up another row with kinase + substrate + ATP as optimized above + 2% DMSO or buffer.
For research use only. Not for use in diagnostic procedures.
The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.




























