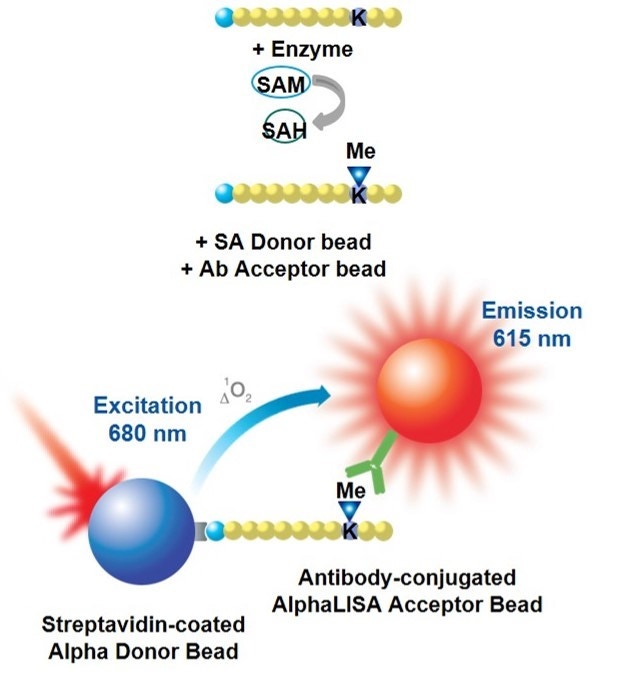
Overview
AlphaLISA™ epigenetics toolbox reagents were designed for the detection of specific methylation and acetylation marks on biotinylated-histone H3 peptide substrates (assay development using a full-length Histone H3 substrate is also feasible). Reagents that detect other post-translational modifications (such as acetylation of p53) are also available. In AlphaLISA assays, a biotinylated histone peptide substrate is used in a biochemical enzymatic reaction with a histone methyltransferase (HMT) or a histone acetyltransferase (HAT) enzyme. After the enzymatic reaction is stopped, the level of methylation or acetylation is determined using antibody-coated AlphaLISA Acceptor beads and streptavidin-coated Alpha Donor beads. The streptavidin Donor beads capture the biotin moiety on the peptide, whereas the antibody on the AlphaLISA Acceptor beads recognizes the specific epigenetic mark (acetylation or methylation). Alternatively, a biotinylated anti-Histone H3 antibody (specific to the C-terminus of Histone H3) can be used to capture full-length Histone H3 substrates on the streptavidin Donor beads. Upon laser irradiation of the bead complexes at 680 nm, short-lived singlet oxygen molecules produced by the Donor beads can reach the Acceptor beads in proximity to generate an amplified chemiluminescent signal at 615 nm. The intensity of light emission is proportional to the level of substrate modification.

Figure 1. Assay principle for AlphaLISA epigenetics toolbox reagents. Schematic representation of an AlphaLISA methyltransferase assay. In both cases, a biotinylated histone H3 peptide substrate is used in conjunction with streptavidin Donor beads and anti-mark antibody-coated AlphaLISA Acceptor beads specific to the particular modification (acetylated or methylated residue).
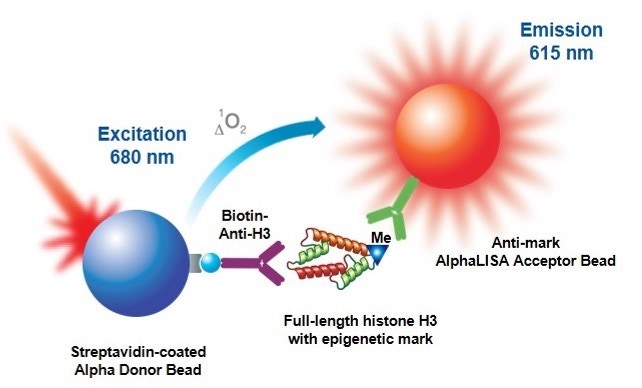
Figure 2. Assay principle using a full-length Histone H3 substrate. A biotinylated antibody specific to the C-terminus of Histone H3 is used to capture the substrate on the streptavidin Donor beads.
It is also possible to perform de-acetylation or de-methylation assays, starting with the appropriate modified substrates that can be detected by the antibody-coated AlphaLISA Acceptor bead and monitoring a decrease in signal as the substrate is de-acetylated or de-methylated.
Interestingly, deacetylase and demethylase assays can also be configured as signal increase assays. This is possible when an anti-non-modified residue antibody is available. Another possibility, only available for demethylase assays, is to use anti-mark antibodies that are specific against mono- or dimethylated lysine residues. With these types of antibodies it will be required to use a di- or trimethylated substrate and to carefully optimize the enzyme concentration and reaction time employed during the enzymatic step to be able to detect the (n-1) methylation state.
Analytical Specificity
Most methylation and acetylation enzymes work by modifying the target substrate at a particular residue or residues. Acetylation occurs on lysine residues, whereas methylation occurs on lysine or arginine residues. Lysine can be mono-, di-, or tri-methylated. Arginine residues can be mono-, and symmetrically or asymmetrically di-methylated.
Currently, AlphaLISA Acceptor beads are offered for these epigenetic and post-translationally-modified targets:
|
Detected modification |
Description of modification |
Catalog number |
|---|---|---|
|
H3K9ac |
Histone H3 acetylated on lysine 9 |
AL114 |
|
H3K27ac |
Histone H3 acetylated on lysine 27 |
AL120 |
|
p53K382ac |
p53 protein acetylated on lysine 382 |
AL124 |
|
H3K4(unmodified) |
Histone H3 unmodified at lysine 4 |
AL119 |
|
H3K4me1-2 |
Histone H3 mono- or di-methylated on lysine 4 |
AL116 |
|
H3K9me2 |
Histone H3 di-methylated on lysine 9 |
AL117 |
|
H3K27me2-1 |
Histone H3 mono- or di-methylated on lysine 27 |
AL121 |
|
H3K27me3 |
Histone H3 tri-methylated on lysine 27 |
AL122 |
|
H3K36me2 |
Histone H3 di-methylated on lysine 36 |
AL123 |
|
H3R2me |
Histone H3 methylated at arginine 2 |
AL139 |
|
H4 acetyl-Lysine (generic) |
Acetylated lysine residues on Histone H4 |
AL143 |
|
O-linked GlcNAc |
O-linked n-acetylglucosamine |
AL144 |
|
H4R3Me |
Histone H4 methylated at arginine3 |
AL150 |
|
H3/H4Rme pan |
Histone H3 and H4 methylated arginine residues |
AL151 |
|
H4K20me1 |
Histone H4 that is mono-methylated at lysine 20 |
AL145 |
|
Histone H3 (C-term) |
Histone H3 in a homogeneous AlphaLISA assay for DOT1L. Use in conjunction with biotinylated anti-H3K79me2 (AL148) |
AL147 |
|
H3K79me2 |
Human Histone H3 that is dimethylated on Lysine 79 |
AL148 |
Analytical specificity of AlphaLISA-conjugated antibodies has been assessed in a detection assay using biotinylated peptides bearing different epigenetic marks. Please see examples in figures 3 to 5, below. Additional information can be obtained by contacting technical support.
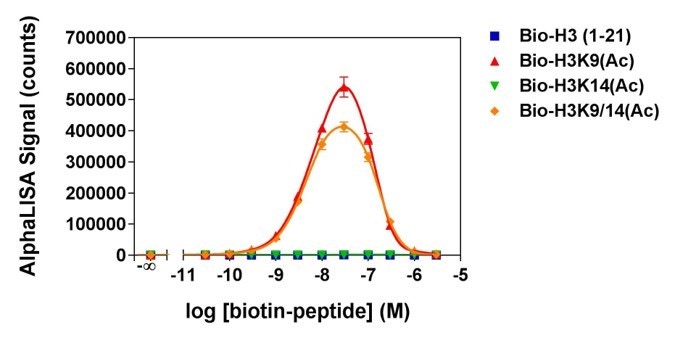
Figure 3. Analytical specificity of AL114, which detects H3K9ac (Histone H3 acetylated lysine 9 peptide). Histone H3-derived peptides with different epigenetic marks were titrated. Signal was detected with an EnVision® HTS Alpha instrument 2102. The hook effect observed at higher peptide concentrations is typical of multi-component assays and occurs when peptide concentrations exceed the binding capacity of the Alpha Donor and/or AlphaLISA Acceptor beads.
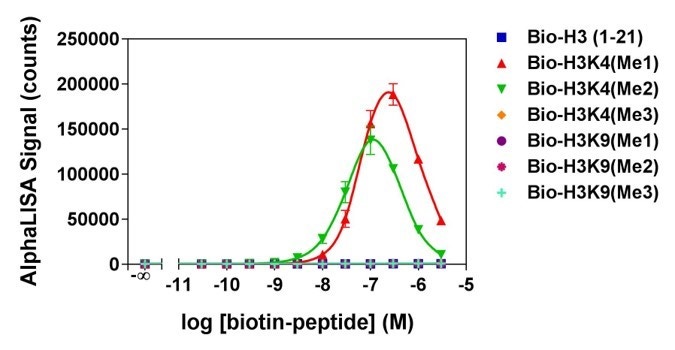
Figure 4. Analytical specificity of AL116, which detects H3K4me1-2 (Histone H3 mono- or di-methylated lysine 4 peptides). Histone H3-derived peptides with different epigenetic marks were titrated. Signal was detected with an EnVision® HTS Alpha instrument 2102. The hook effect observed at higher peptide concentrations is typical of multi-component assays and occurs when peptide concentrations exceed the binding capacity of the Alpha Donor and/or AlphaLISA Acceptor beads.
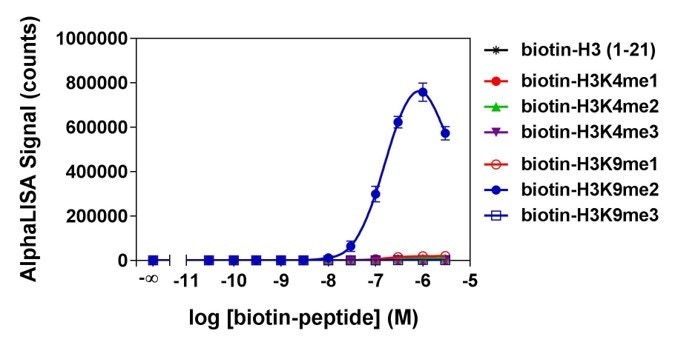
Figure 5. Analytical specificity of AL117C, which detects H3K9me2 (Histone H3 di-methylated lysine 9 peptide). Histone H3-derived peptides with different epigenetic marks were titrated. Signal was detected with an EnVision™ HTS Alpha instrument 2102. The hook effect observed at higher peptide concentrations is typical of multi-component assays and occurs when peptide concentrations exceed the binding capacity of the Alpha Donor and/or AlphaLISA Acceptor beads.
Custom beads may also be available upon request, and custom assay development services are offered through our OnPoint custom teams (contact information is at the bottom of this page). It is also possible to use our unconjugated beads and other bead products to develop your own assay.
What do I need to run this assay?
Reagents available from Revvity:
- Antibody-coated AlphaLISA Acceptor beads, specific to an acetylation or methylation site (refer to Products and Catalog numbers section below, or table in section above)
- Streptavidin-coated Donor beads - required only if using biotinylated Histone peptide substrates
- Biotin anti-Histone H3 antibody (C-terminal specific - Cat. No. AL118) – required only if you are developing an assay using a full-length Histone H3 substrate, rather than a biotinylated peptide substrate
- Biotin anti-Histone H4 antibody (N-terminal specific - Cat. No. AL146) - required only if you are developing an assay using full-length Histone H4 substrate, rather than a biotinylated peptide substrate
- Microplates (We recommend our white OptiPlate™ microplates or our light grey AlphaPlate™ microplates - refer to Microplate selection for AlphaLISA assays for more information.)
- TopSeal-A™ adhesive seal to prevent evaporation
- Plate reader capable of measuring Alpha assays (We recommend our EnVision™ Multilabel Plate Reader or VICTOR Nivo™ Multilabel Plate Reader.)
Reagents available from AnaSpec or other suppliers:
- Biotinylated Histone H3 peptide or p53 peptide substrate. Depending on your enzyme, you may need to start with a non-modified substrate, or you may need to start with a substrate that is otherwise pre-modified at a specific position. In general, we recommend using a non-modified (bare) peptide if you are performing acetylation or methylation assays. In any case, make sure your assay is designed so that the antibody on the AlphaLISA Acceptor bead differentiates between substrate and product. If you are trying a deacetylation or demethylation assay, you will need to start with a substrate that is pre-modified (for example, di-methylated on lysine 9, acetylated on lysine 9, etc.).
- Additionally, you may wish to purchase a second peptide to be used as a positive control for your assay. For example, if you are measuring acetylation of lysine 9 on Histone H3 peptide, you may wish to purchase the biotinylated acetyl-Lys9 Histone H3 peptide.
| Product | Sequence | AnaSpec catalog number |
|---|---|---|
| Biotinylated Histone H3 peptide (residues 1-21), non-modified (bare) | ARTKQTARKSTGGKAPRKQLA-GGK(biotin)-Nh3 | #61702 |
| Biotinylated Histone H3 peptide (residues 21-44), non-modified (bare) | ATKAARKSAPATGGVKKPHRYRPG-GK(biotin) | #64440 |
| Biotinylated Histone H3 peptide, acetylated on Lysine 9 | ARTKQTAR-K(Ac)-STGGKAPRKQLA-GGK(biotin)-Nh3 | #64361 |
| Biotinylated Histone H3 peptide, acetylated on Lysine 14 | ARTKQTARKSTGG-K(Ac)-APRKQLA-GGK(biotin)-Nh3 | #64362 |
| Biotinylated Histone H3 peptide, acetylated on Lysine 9 and Lysine 14 | ARTKQTAR-K(Ac)-STGG-K(Ac)-APRKQLA-GGK-biotin-Nh3 | #64363 |
| Biotinylated Histone H3 peptide, acetylated on Lysine 27 | ATKAAR-K(Ac)-SAPATGGVKKPHRYRPGG-K(biotin) | #64846 |
| Biotinylated Histone H3 peptide, mono-methylated on Lysine 4 | ART-K(Me1)-QTARKSTGGKAPRKQLA-GGK-biotin-Nh3 | #64355 |
| Biotinylated Histone H3 peptide, di-methylated on Lysine 4 | ART-K(Me2)-QTARKSTGGKAPRKQLA-GGK(biotin)-Nh3 | #64356 |
| Biotinylated Histone H3 peptide, tri-methylated on Lysine 4 | ART-K(Me3)-QTARKSTGGKAPRKQLA-GGK(biotin)-Nh3 | #64357 |
| Biotinylated Histone H3 peptide, mono-methylated on Lysine 9 | ARTKQTAR-K(Me1)-STGGKAPRKQLA-GGK(biotin)-Nh3 | #64358 |
| Biotinylated Histone H3 peptide, di-methylated on Lysine 9 | ARTKQTAR-K(Me2)-STGGKAPRKQLA-GGK(biotin)-Nh3 | #64359 |
| Biotinylated Histone H3 peptide, tri-methylated on Lysine 9 | ARTKQTAR-K(Me3)-STGGKAPRKQLA-GGK(biotin)-Nh3 | #64360 |
| Biotinylated Histone H3 peptide, mono-methylated on Lysine 27 | ATKAAR-K(Me1)-SAPATGGVKKPHRYRPG-GK(biotin) | #64365 |
| Biotinylated Histone H3 peptide, di-methylated on Lysine 27 | ATKAAR-K(Me2)-SAPATGGVKKPHRYRPG-GK(biotin) | #64366 |
| Biotinylated Histone H3 peptide, tri-methylated on Lysine 27 | ATKAAR-K(Me3)-SAPATGGVKKPHRYRPG-GK(biotin) | #64367 |
| Biotinylated Histone H3 peptide, di-methylated on Lysine 36 | ATKAARKSAPATGGV-K(Me2)-KPHRYRPG-GK(biotin) | #64442 |
| Biotinylated Histone H3 peptide, tri-methylated on Lysine 36 | ATKAARKSAPATGGV-K(Me3)-KPHRYRPG-GK(biotin) | #64441 |
| Biotinylated p53 peptide (residues 361-393), non-modified (bare) | Biotin-LC-GSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD-Nh3 | #62530 |
| Biotinylated p53 peptide, acetylated on Lysine 382 | Biotin-KGGHLKSKKGQSTSRHK-K(Ac)-LMFKTEGPDSD-Nh3 | #64869 |
Full-length recombinant Histone H3 substrate (Active Motif Cat. No. 31207), if you would like to develop an assay using a full-length Histone H3 protein rather than a biotinylated peptide substrate.
Reagents available from various suppliers:
- Cofactor (acetyl CoA for acetylation assays, or S-adenosylmethionine for methylation assays - see Tips and FAQs section below to see where Revvity R&D gets these reagents.)
- Enzyme
- Reaction buffer
- Test compounds as desired
Protocol-in brief
The assay is performed in two steps: first, the enzymatic assay is performed in an appropriate reaction buffer. Then, the reaction is stopped and Acceptor and Donor beads are added sequentially in 1X AlphaLISA Epigenetics Buffer 1 for the detection step.
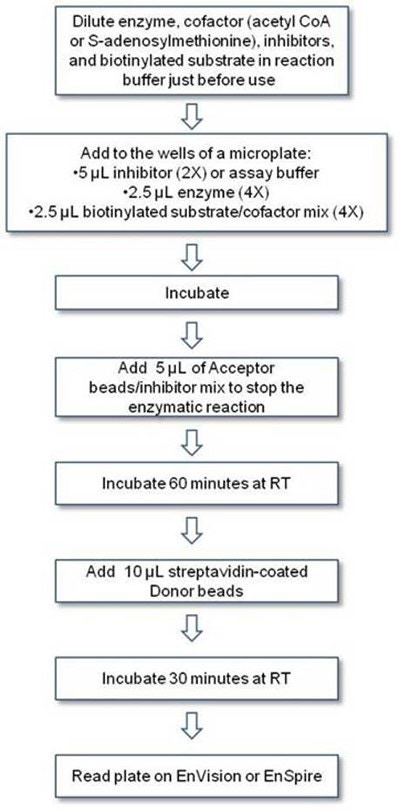
Figure. Alpha epigenetic workflow
Assay development
Assays have to be optimized using your particular enzyme/substrate pair. Examples of assay development for a number of enzymes can be found in the format of Technical Notes. The example below describes the optimization of a p300 acetyltransferase assay using anti-H3K9ac Acceptor Beads (as found in the technical note).
1. Enzyme titration and time-course.
This experiment is set up using increasing concentrations of enzyme where multiple wells are filled for each enzyme concentration allowing stopping the reaction at different time points. For subsequent assay optimization, you will want to choose a time and enzyme concentration providing for a linear increase of reaction product over time, conserving enzyme while still achieving good signal.
Keep in mind that if you are performing a methylation assay, stopping the reaction at the appropriate time could be critical as several of our LANCE Europium antibodies detect specific methylation states. For example, if a methylation reaction were to proceed for too long a time, substrate tri-methylation could occur (provided the enzyme used had that capacity), with an eventual loss of signal if the antibody was specific for the di-methylated residue.
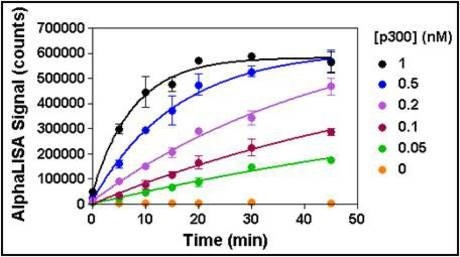
Figure 6. Enzymatic progress curves were performed by incubating p300 at concentrations ranging from 0.05 to 1 nM with 50 nM biotinylated H3 (1-21) peptide substrate and 25 µM acetyl CoA. A mix of Acceptor beads and anacardic acid was added to stop the reactions at the indicated times. Donor beads were added 60 min later and signal was read after 30 min. A 15-minute reaction time using 0.2 nM enzyme was selected for all subsequent experiments.
2. Cofactor titration.
This assay is performed to determine the apparent Km for the cofactor (either acetyl CoA for acetylation assays, or S-adenosylmethionine for methylation assays). Increasing concentrations of cofactor are tested at the enzyme concentration and time point chosen from the previous experiment. For most assays, you will want to choose a concentration of cofactor that is at or close to the apparent Km.
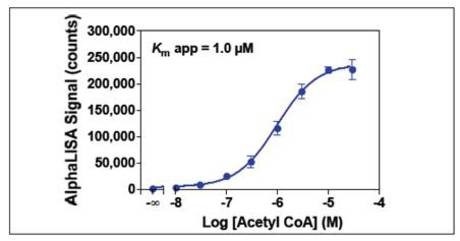
Figure 7. Serial dilutions of acetyl CoA ranging from 10 nM to 30 µM were added to 0.2 nM p300 and 50 nM biotinylated H3 (1-21) peptide substrate. A 3 µM acetyl CoA concentration was selected for subsequent experiments.
3. Enzyme inhibition.
This assay can be used to characterize inhibitors for your enzyme, or as part of assay validation using a reference inhibitor if you will be screening. Increasing concentrations of inhibitor are tested at the enzyme concentration, time point, and cofactor concentration determined from the previous two experiments. An IC50 for the inhibitor can be derived from this data. It is important to note that if the compounds to be tested are dissolved in a solvent that is different than water (typically DMSO), its effect on the enzymatic reaction must be evaluated beforehand.

Figure 8. Serial dilutions of anacardic acid ranging from 300 nM to 300 µM and garcinol ranging from 200 nM to 200 µM were pre-incubated for 10 min with 0.2 nM p300. Enzymatic reactions were initiated by the addition of 50 nM biotinylated H3 (1-21) peptide substrate plus 3 μM acetyl CoA. Enzymatic reactions contained 1% DMSO.
4. Z'-factor determination.
If screening, you may wish to perform a Z'-factor determination experiment as part of your assay validation. Two conditions are chosen that will generate a high and low signal (for example, with and without a particular concentration of inhibitor, or with and without a particular concentration of cofactor). In general, a Z'-factor above 0.5 is highly desirable for screening campaigns.
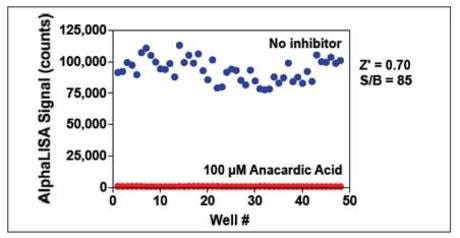
Figure 9. p300 (0.2 nM) was pre-incubated with or without 100 µM anacardic acid for 10 min. Enzymatic reactions were initiated by the addition of 50 nM biotinylated H3 (1-21) peptide substrate plus 3 μM acetyl CoA. Enzymatic reactions contained 1% DMSO.
Tips and FAQs
- Acetyltransferase and methyltransferase recombinant enzymes can exhibit low activity in in vitro biochemical assays, compared to other types of enzymes such as kinases. Reaction buffer optimization (including pH, salt, surfactant concentration, etc.), enzyme titration, and time-course experiments are critical in achieving good signal window.
- In order to avoid stability issues and minimize assay variability, enzyme, cofactor/s, substrate/s, and test compounds should be diluted in assay buffer just before use.
- Acetyl CoA cofactor is prepared at 50 mg/mL (56.9 mM) in h3O, aliquoted and stored at -80 °C. Revvity R&D obtains acetyl CoA from Sigma (#A2056).
- S-adenosylmethionine is prepared at 30 mM in 5 mM h3SO4/10% ethanol (v/v) in h3O, aliquoted, and stored at -80 °C. Revvity R&D obtains S-adenosylmethionine from Sigma (#A7007).
- AlphaLISA 5X Epigenetics Buffer 1 Kit comes as a two-component buffer kit containing the AlphaLISA 5X Buffer 1 plus a 30X Epigenetics Buffer Supplement, which are used to prepare the 1X AlphaLISA Epigenetics Buffer 1. The buffer prevents non-specific binding of highly-charged histone peptides to the Acceptor and Donor beads in AlphaLISA Epigenetic assays. It was specifically formulated for AlphaLISA reagents detecting Histone H3 (1-21) peptide acetylation and methylation (AL114, AL116, AL117). The 1X AlphaLISA Epigenetics Buffer 1 is to be used at the Detection step for the dilution of AlphaLISA Acceptor and Alpha Donor beads and is not intended as an assay buffer for the enzymatic reaction step. This buffer is not recommended for the detection of full-length histone proteins or nucleosomes.
- The concentrations of biotin-peptide substrate indicated in the Technical Notes are the best concentrations to work with for the model enzymes indicated. For other enzymes, or if you would like to use a different peptide (longer, shorter, bearing other modifications already, etc.), it is possible that you will need to adjust the substrate concentration. Nevertheless, we suggest that you initially use the substrate concentration indicated on the technical notes (for a given anti-mark antibody) to generate your enzyme progression curves. You should only start adjusting the biotin-peptide substrate concentration if the enzymatic assay does not work well and other components in the assay have been optimized already. If possible, we strongly advise that you try changing the concentration of the co-factor or enzyme, or the reaction time rather than changing the concentration of biotin-peptide substrate. An important thing to keep in mind is that, generally, the concentration of biotin-peptide substrate cannot be increased too much because this could cause the detection assay to “hook” on the biotin/streptavidin binding site.
For research use only. Not for use in diagnostic procedures.
The information provided above is solely for informational and research purposes only. Revvity assumes no liability or responsibility for any injuries, losses, or damages resulting from the use or misuse of the provided information, and Revvity assumes no liability for any outcomes resulting from the use or misuse of any recommendations. The information is provided on an "as is" basis without warranties of any kind. Users are responsible for determining the suitability of any recommendations for the user’s particular research. Any recommendations provided by Revvity should not be considered a substitute for a user’s own professional judgment.