Introduction
Long non-coding RNAs (lncRNAs) represent a substantial and functionally diverse component of the transcriptome, implicated in the regulation of gene expression and cellular processes across various biological contexts. Building on prior discussions of lncRNA architecture and expression , this blog focuses on functional genomics approaches that extend beyond transcript identification to elucidate the roles of lncRNAs at a genome-wide scale.
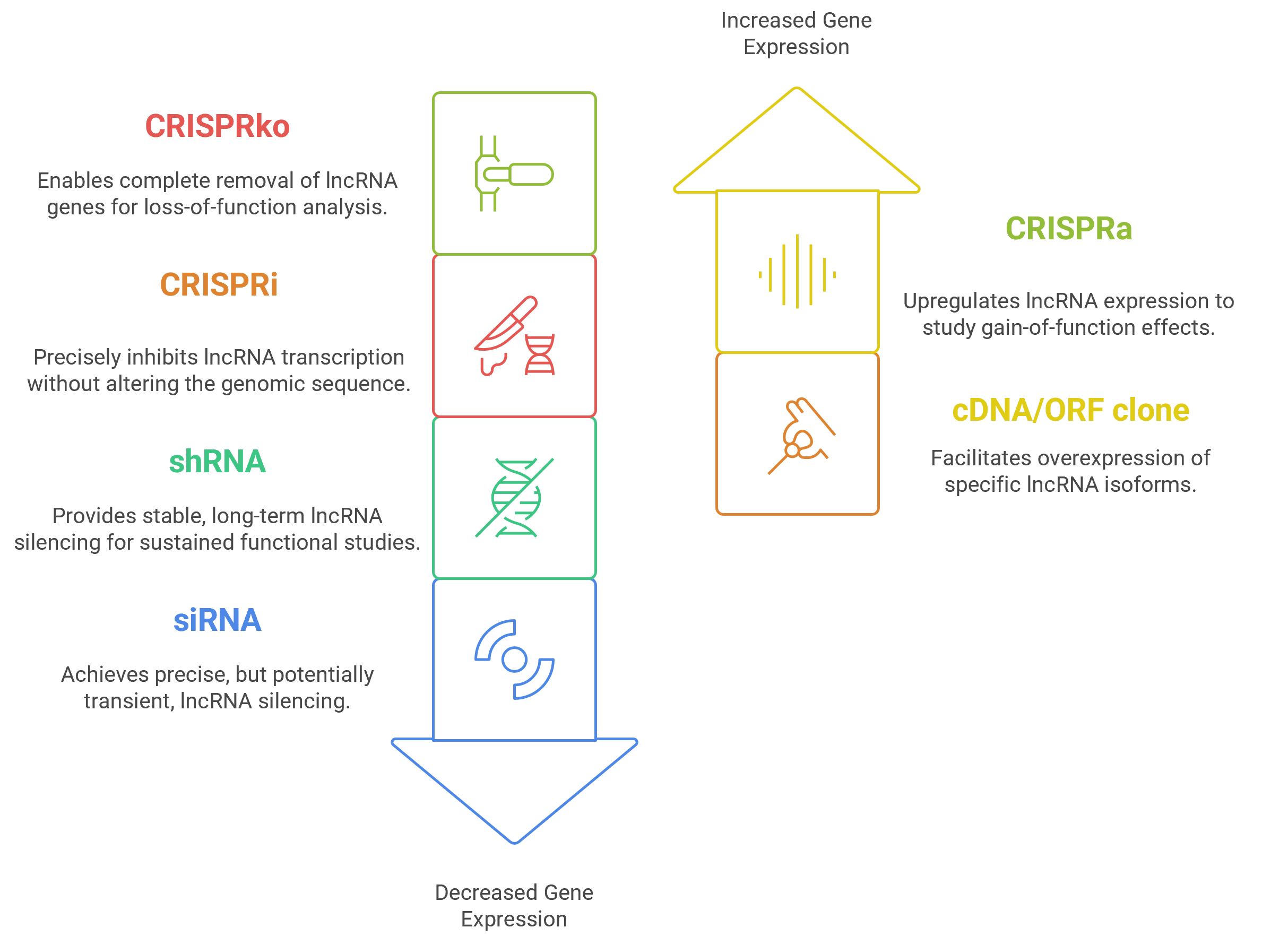
CRISPR-based screening methods have transformed the study of lncRNAs by allowing accurate and scalable adjustments to their expression. Alongside RNA interference techniques and overexpression systems, these tools can confirm lncRNA roles in cellular models with exceptional clarity. These orthogonal approaches are crucial for deepening our knowledge of lncRNA biology and speeding up their development into biomarkers and therapeutic targets. By combining high-resolution sequencing with functional perturbation techniques, researchers can dissect the regulatory networks orchestrated by lncRNAs, shedding light on their roles in cancer, neurodegeneration, and immune regulation
For research use only. Not for diagnostic purposes.
Genome-Scale Technologies for lncRNA functional studies
LncRNA Discovery
With broad dynamic range and high sensitivity, RNA sequencing
 NEXTFLEX Rapid Directional RNA-Seq Kit 2.0
Discover
remains the cornerstone for discovering and quantifying lncRNAs. RNA-seq allows researchers profiling the entire transcriptome without relying on pre-designed probes or prior knowledge of gene annotations. RNA-seq can also identify alternatively spliced transcripts of lncRNAs, revealing the complexity and diversity of lncRNA species that may have distinct functions.
NEXTFLEX Rapid Directional RNA-Seq Kit 2.0
Discover
remains the cornerstone for discovering and quantifying lncRNAs. RNA-seq allows researchers profiling the entire transcriptome without relying on pre-designed probes or prior knowledge of gene annotations. RNA-seq can also identify alternatively spliced transcripts of lncRNAs, revealing the complexity and diversity of lncRNA species that may have distinct functions.
Spatial Mapping
Chromatin Isolation by RNA Purification followed by sequencing (ChIRP-seq) is a powerful technique used to map the genome-wide binding sites of lncRNAs. ChIRP-seq has uncovered that lncRNA binding sites are often focal, sequence-specific, and numerous, resembling transcription factor occupancy rather than broad histone modification patterns.
Overexpression /Repression
cDNA and ORF
cDNA and open reading frame (ORF) clone libraries facilitate overexpression of specific lncRNA isoforms, allowing functional dissection of domain-specific and variant-specific effects on cellular phenotypes. Some of the cDNA clones available in the Mammalian Gene Collection target lncRNAs.
CRISPR technologies
Effective modulation of lncRNAs using CRISPR technology depends on the careful design of custom single guide RNAs with high specificity and efficiency. We have developed a dedicated sgRNA design platform (Dharmacon™ CRISPR Design Tool ) that integrates a user-friendly interface with sophisticated computational methods to identify optimal target sequences within gene bodies. This platform incorporates predictive models for off-target interactions and evaluates sgRNA activity to support reliable CRISPR knockout (KO) experiments. For example, CRISPR can be used to excise a lncRNA gene using 2 guides flanking it, to generate a knockout cell line.
Beyond conventional gene knockout, the development of CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) has unlocked the ability to finely tune lncRNA expression without altering genomic sequences
- CRISPRi employs a nuclease-deactivated Cas9 (dCas9) fused to repressive domains to inhibit transcription initiation by targeting promoters or enhancers.
- CRISPRa uses dCas9 fused to activator domains to upregulate lncRNA expression in a physiological context.
Our in-house RNA synthesis capabilities enable the production of high-quality custom reagents to support CRISPR interference and activation studies
Precision Knockdown with Optimized siRNA
siRNA research tools incorporating chemical modifications, such as Lincode™ siRNA, reduce off-target effects by inactivating passenger strand activity and minimizing seed region miRNA-like effects enhancing specificity and knockdown efficiency. These same tools can be synthesized with modifications that allow for self-delivery into cells, and/or, for in-vivo nuclease resistance enabling animal models. In breast cancer models, siRNA-mediated silencing of the immunomodulatory lncRNA LINK-A disrupted its interaction with HIF1α, restoring T-cell-mediated antitumor immunity and illustrating the therapeutic potential of targeted lncRNA knockdown.
Sustained knockdown using the SMARTvector platforms
SMARTvector™ lentiviral shRNA, developed using our advanced microRNA-based shRNA rational design algorithm and expressed under an optimized promoter, can be used for highly potent and specific lncRNA gene silencing. For applications requiring temporal control, SMARTvector inducible reagents enable the rapid generation of stable cell lines with tightly regulated shRNA expression. The Tet-On 3G promoter system provides minimal basal expression and strong inducibility, allowing near wild-type levels of target gene expression in the absence of doxycycline and robust gene knockdown upon induction.

Computational Integration and Biomarker Discovery
Machine learning classifiers like LncDC accurately distinguish lncRNAs from mRNAs by analyzing sequence motifs and secondary structures, aiding annotation in large datasets. Databases such as lnc2Cancer 3.0 curate experimentally validated lncRNA-disease associations, helping prioritize candidates for functional validation.
Conclusion: toward lncRNA-Driven Therapeutics
The integration of advanced sequencing, functional genomics, and RNAi technologies is changing our understanding of lncRNAs. Dharmacon Lincode™ siRNA, SMARTvector shRNA technology, and the sgRNA design platform tools exemplify how tailored, high-specificity reagents can overcome historical challenges in targeting lncRNAs. Coupled with computational prediction and biomarker databases, these platforms enable the translation of lncRNA biology into novel diagnostics and therapeutics.
By linking the structural and sequencing insights from our previous blog with the functional perturbation strategies discussed here, researchers are equipped with a powerful, multi-dimensional toolkit to decode lncRNA regulatory networks and unlock their full potential.
References:
- Mattick, J.S., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nature Reviews Molecular Cell Biology, 24, 430–447.
- Xu, J., et al (2025). Identification of long noncoding RNAs (lncRNAs) and co-transcriptional analysis of mRNAs and lncRNAs in transcriptomes of Anopheles gambiae Frontiers in RNA Research, 3.
- Zare, K, et al (2018). CRISPR/Cas9 Knockout Strategies to Ablate CCAT1 lncRNA Gene in Cancer Cells. Biol Proced Online. Nov 1;20:21
- Zibitt M.S., et al (2021). Interrogating lncRNA functions via CRISPR/Cas systems. RNA Biol. 18(12):2097-2106.
- Liu, S.J. et al. (2017). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355,eaah7111.
- Bester, A.C. et al (2018). An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell. 173(3) 649-664.e20
For research use only. Not for use in diagnostic procedures.